Imatinib
Overview
Description
Imatinib (marketed as Gleevec/Glivec) is a first-generation tyrosine kinase inhibitor (TKI) developed for the treatment of chronic myeloid leukemia (CML) and gastrointestinal stromal tumors (GISTs). It selectively targets the Bcr-Abl fusion protein, a constitutively active tyrosine kinase responsible for uncontrolled proliferation in CML . This compound also inhibits platelet-derived growth factor receptor (PDGFR) and c-Kit kinases, broadening its therapeutic applications .
Preparation Methods
The synthesis of Imatinib involves a classical convergent approach. The compound is assembled by coupling the amine and carboxylic acid precursors using N,N′-carbonyldiimidazole (CDI) as a condensing agent . The first synthesis of this compound, reported by Zimmermann, was based on the connection of two key intermediates: the aromatic amine and 4-[(4-methylpiperazin-1-yl)methyl]benzoyl chloride . Industrial production methods have been optimized to overcome certain problematic steps, save time and labor, and provide high yield and purity .
Chemical Reactions Analysis
Imatinib undergoes various types of chemical reactions, including oxidation, reduction, and substitution. Common reagents and conditions used in these reactions include hydrogen over Adam’s catalyst for reduction and CDI for condensation reactions . Major products formed from these reactions include various this compound analogues and impurities, which are characterized and confirmed using techniques such as IR, 1H-NMR, and Mass Spectrometry .
Scientific Research Applications
Indications for Imatinib
This compound is primarily indicated for the following conditions:
- Chronic Myeloid Leukemia (CML) : Effective in all phases, particularly in patients with the Philadelphia chromosome.
- Acute Lymphoblastic Leukemia (ALL) : Specifically for Ph+ ALL in adults and children.
- Gastrointestinal Stromal Tumors (GIST) : Particularly those expressing c-KIT mutations.
- Aggressive Systemic Mastocytosis : Without D816V c-KIT mutations.
- Hypereosinophilic Syndrome (HES) : Especially with FIP1L1-PDGFR-alpha fusion kinase.
- Dermatofibrosarcoma Protuberans : In unresectable cases.
Chronic Myeloid Leukemia (CML)
This compound has been extensively studied in CML, showing significant efficacy. The IRIS trial demonstrated a 10-year overall survival rate of 83.3% in patients treated with this compound as first-line therapy .
| Phase | Major Cytogenetic Response (%) | Complete Hematologic Response (%) |
|---|---|---|
| Chronic Phase | 60 | 95 |
| Accelerated Phase | 24 | 82 |
| Blast Crisis | 16 | 52 |
Acute Lymphoblastic Leukemia (ALL)
In Ph+ ALL, this compound combined with chemotherapy resulted in a 5-year disease-free survival rate of 70% compared to 65% with sibling donor bone marrow transplants .
Gastrointestinal Stromal Tumors (GIST)
This compound has shown remarkable effectiveness in GIST treatment. In randomized phase III trials, patients on a daily dose of 400 mg achieved a median progression-free survival of approximately 18.9 months .
| Study Type | Response Rate (%) | Overall Survival (months) |
|---|---|---|
| Phase III Trial | 83 | 49 |
Other Applications
This compound's application extends to several other malignancies:
- Aggressive Systemic Mastocytosis : Achieved complete hematologic response in about 29% of patients .
- Hypereosinophilic Syndrome : Induced complete hematologic response in approximately 61% of cases .
- Dermatofibrosarcoma Protuberans : An overall response rate of 83%, with a complete response rate of 39% reported .
Mechanism of Action
Imatinib works by inhibiting the Bcr-Abl tyrosine kinase, the constitutive abnormal gene product of the Philadelphia chromosome in chronic myeloid leukemia (CML) . Inhibition of this enzyme blocks proliferation and induces apoptosis in Bcr-Abl positive cell lines as well as in fresh leukemic cells in Philadelphia chromosome-positive CML . This mechanism helps stop the spread of cancer cells .
Comparison with Similar Compounds
Mechanism of Action
Imatinib binds to the ATP-binding pocket of Bcr-Abl, stabilizing the kinase in an inactive conformation (DFG-out state), thereby blocking downstream signaling pathways critical for leukemogenesis . Structural studies show that its methylbenzene group and hydrogen-bonding interactions with residues like Glu286 and Ala381 are pivotal for binding affinity .
Clinical Efficacy
The IRIS trial (6-year follow-up) demonstrated this compound’s long-term efficacy in CML chronic phase (CML-CP):
- Complete cytogenetic response (CCyR) rate : 82%
- Overall survival (OS): 88% (95% when excluding non-CML-related deaths)
- Event-free survival (EFS) : 83% .
In a Chinese cohort of 54 CML patients, this compound achieved 94.1% hematologic remission in CP patients, though responses diminished in advanced phases (61.5% in accelerated phase; 37.5% in blast crisis) .
Pharmacokinetics and Metabolism
This compound is primarily metabolized by CYP3A4 into its active metabolite, N-desmethyl this compound. Drug-drug interactions with CYP3A4 inducers (e.g., isoflavones) can reduce this compound’s plasma concentration, necessitating dose adjustments .
Structural and Binding Profile Comparisons
Nilotinib
- Structure : Optimized from this compound by reversing the amide orientation, enhancing binding to the Abl kinase domain .
- Potency : 30-fold greater affinity for Bcr-Abl than this compound due to improved hydrophobic interactions and hydrogen bonding .
- Clinical Impact : Effective in this compound-resistant CML, particularly against mutations like T315I (except in ponatinib-sensitive cases) .
Dasatinib
- Structure : Binds Abl in both active and inactive conformations, enabling inhibition of multiple Bcr-Abl mutants .
- Multitargeting : Inhibits Src kinases, broadening activity but increasing off-target toxicity (e.g., pleural effusions) .
SKI696 (this compound Analog)
- Design : Radiolabeled analog retaining this compound’s kinase selectivity (PDGFRα, c-Kit, Abl) but modified for imaging applications.
- Inhibition Profile: Kinase this compound (100 nM) SKI696 (100 nM) PDGFRα 100% 78.2% c-Kit 30.6% 30.6% Abl 33.4% 10.4% Minor differences arise from structural modifications for radiolabeling .
Efficacy in Resistance Mutations
- T674M (PDGFRα) : Both this compound and nilotinib show reduced efficacy, but compound 11 (flag-methyl-modified) maintains activity .
- Kit-T670I : this compound derivatives lacking N-methylpiperazine (e.g., compound 5) exhibit diminished potency, highlighting the substituent’s role in solubility and target engagement .
Toxicity and Selectivity
- This compound: Lower rodent carcinogenicity compared to newer TKIs but associated with developmental toxicity in preclinical models .
- Novel PDGFRA Inhibitors: Computational studies identified 13 compounds with higher intestinal absorption and reduced mutagenicity vs. This compound, though none matched its kinase specificity .
Polypharmacology and Repurposing Potential
It interacts with fewer proteins (predicted: 15–20) than broad-spectrum inhibitors like staurosporine, minimizing off-target effects .
Biological Activity
Imatinib, a selective tyrosine kinase inhibitor (TKI), has transformed the management of various malignancies, particularly chronic myeloid leukemia (CML) and gastrointestinal stromal tumors (GISTs). This article delves into the biological activity of this compound, highlighting its mechanisms of action, clinical efficacy, and relevant case studies.
This compound primarily targets the BCR-ABL fusion protein, a product of the Philadelphia chromosome, which is a hallmark of CML. By inhibiting the tyrosine kinase activity associated with BCR-ABL, this compound effectively disrupts downstream signaling pathways that promote cell proliferation and survival. The compound binds to the ATP-binding site of the kinase, stabilizing it in an inactive conformation and preventing phosphorylation of tyrosine residues on substrate proteins, thereby inducing apoptosis in malignant cells .
Key Mechanisms:
- Inhibition of Tyrosine Kinases: this compound inhibits several tyrosine kinases including ABL, PDGFRA, and c-KIT.
- Induction of Apoptosis: The inhibition of BCR-ABL leads to reduced anti-apoptotic signaling, promoting cell death in affected leukemic cells.
- Selective Activity: this compound exhibits selective cytotoxicity against BCR-ABL-positive cells while sparing normal cells that utilize other redundant pathways .
Clinical Efficacy
This compound's clinical efficacy has been extensively documented across multiple studies. Below is a summary table showcasing key clinical findings:
Case Studies and Research Findings
-
Chronic Myeloid Leukemia (CML):
A landmark study demonstrated that patients with CML treated with this compound achieved unprecedented rates of complete cytogenetic response (CCR), with over 80% success in newly diagnosed cases. Resistance often emerged due to mutations in the BCR-ABL kinase domain . -
Gastrointestinal Stromal Tumors (GIST):
A recent trial emphasized the importance of uninterrupted this compound therapy for advanced GISTs. Patients who continued treatment showed significant improvements in survival compared to those who discontinued therapy . -
COVID-19 Implications:
Emerging research indicates potential benefits of this compound in severe COVID-19 cases. A randomized trial suggested that this compound may reduce mortality and mechanical ventilation duration in critically ill patients, warranting further investigation into its role beyond oncology .
Adverse Effects and Resistance
While this compound is generally well-tolerated, adverse effects can include gastrointestinal disturbances, edema, and hematological changes. Resistance mechanisms often involve mutations in the BCR-ABL gene that alter the binding affinity of this compound. These mutations necessitate alternative therapeutic strategies or second-generation TKIs for effective management .
Q & A
Basic Research Questions
Q. How should researchers design preclinical experiments to evaluate Imatinib’s efficacy in chronic myeloid leukemia (CML)?
- Methodological Answer : Begin with in vitro dose-response assays using BCR-ABL+ cell lines, referencing prior studies that established IC50 values (e.g., 0.25–1.0 µM) . Validate findings with in vivo models (e.g., xenografts) using doses of 50–100 mg/kg/day, adjusted for bioavailability . Include controls for off-target effects (e.g., ABL-negative cell lines) and replicate experiments at least three times to ensure statistical power . For translational relevance, cross-reference clinical trial data on plasma trough levels (e.g., ≥1,000 ng/mL correlates with cytogenetic response) .
Q. What factors should be controlled to ensure reliable this compound response data in cell-based assays?
- Methodological Answer : Control for P-glycoprotein (P-gp) expression, which mediates this compound efflux and reduces intracellular drug accumulation . Use flow cytometry to quantify P-gp levels in cell lines and correlate with IC50 shifts. Standardize culture conditions (e.g., serum concentration, pH) to minimize variability in proliferation rates. Include a positive control (e.g., STI571-resistant cell lines with KIT mutations) and validate results with secondary assays (e.g., apoptosis via Annexin V staining) .
Q. How can researchers optimize this compound dosing in early-phase clinical trials?
- Methodological Answer : Use pharmacokinetic/pharmacodynamic (PK/PD) modeling to link plasma trough levels (target: 1,000–3,200 ng/mL) to clinical outcomes . Adjust doses based on patient-specific factors (e.g., hepatic function, drug-drug interactions) and monitor adverse events (e.g., edema, myelosuppression) . For dose escalation, follow phase I protocols with cohorts receiving 25–1,000 mg/day, prioritizing safety and response rates .
Advanced Research Questions
Q. How should researchers analyze contradictory data on this compound’s efficacy in non-CML malignancies (e.g., gastrointestinal stromal tumors [GIST])?
- Methodological Answer : Conduct subgroup analyses stratified by tumor genotype (e.g., KIT exon 11 vs. PDGFRA mutations) . Use multivariate Cox regression to identify confounding variables (e.g., mitotic index ≤5/50 HPFs correlates with prolonged progression-free survival) . Cross-validate findings with independent datasets (e.g., NCT00075426 trial data) and employ sensitivity analyses to assess robustness against outliers .
Q. What experimental strategies can elucidate mechanisms of this compound resistance in advanced CML?
- Methodological Answer : Perform Sanger sequencing of BCR-ABL kinase domains to detect resistance-conferring mutations (e.g., T315I) . Combine in vitro mutagenesis screens with structural modeling to predict mutation impact on drug binding . Validate findings using patient-derived xenografts (PDXs) and correlate with clinical resistance timelines. Explore adjunct therapies (e.g., dasatinib for T315I mutations) .
Q. How can circulating tumor DNA (ctDNA) be utilized to assess minimal residual disease (MRD) in GIST patients discontinuing this compound?
- Methodological Answer : Design longitudinal studies with serial ctDNA sampling during this compound interruption. Use digital PCR or NGS to detect KIT/PDGFRA mutations, with a sensitivity threshold of 0.1% variant allele frequency . Correlate ctDNA dynamics with radiographic progression (RECIST 1.1) and survival outcomes. Note: Current ctDNA platforms may miss non-KIT/PDGFRA mutations, necessitating orthogonal validation (e.g., ddPCR) .
Q. What statistical methods are appropriate for analyzing survival outcomes in this compound interruption trials?
- Methodological Answer : Apply Kaplan-Meier estimates for progression-free survival (PFS) and log-rank tests to compare groups (e.g., complete vs. incomplete tumor resection) . Use Cox proportional hazards models to adjust for covariates (e.g., peritoneal metastasis status, mitotic index) . For small cohorts, employ bootstrapping to estimate confidence intervals and address censoring biases .
Q. Methodological and Ethical Considerations
Q. How should researchers address retracted or conflicting studies in meta-analyses of this compound data?
- Methodological Answer : Exclude retracted papers (e.g., ) after verifying retraction notices in databases like Retraction Watch. Use GRADE criteria to assess study quality and heterogeneity (e.g., I² statistic). For conflicting results, perform sensitivity analyses by excluding outlier studies and report funnel plots to detect publication bias .
Q. What guidelines should govern the compilation of this compound data in Investigator’s Brochures (IBs) for clinical trials?
- Methodological Answer : Follow ICH E6(R2) guidelines: Include pharmacokinetic data (e.g., Cmax, AUC), toxicity profiles, and response rates from phase I-III trials . For novel formulations (e.g., generics), provide bioequivalence data vs. the reference product. Update IBs annually or after significant safety events (e.g., new black-box warnings) .
Q. How can researchers ethically design trials involving this compound interruption in stable GIST patients?
- Methodological Answer :
Obtain informed consent detailing risks of disease progression (61% in 19.6 months) and benefits of re-introduction (88.6% response rate) . Use a Data Safety Monitoring Board (DSMB) to review interim analyses. Ensure rescue protocols (e.g., this compound re-initiation at 400 mg/day) are predefined in the trial protocol .
Properties
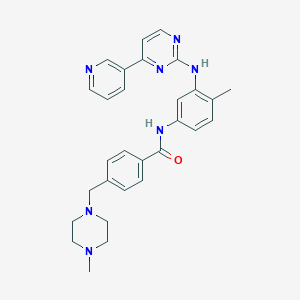
IUPAC Name |
4-[(4-methylpiperazin-1-yl)methyl]-N-[4-methyl-3-[(4-pyridin-3-ylpyrimidin-2-yl)amino]phenyl]benzamide | |
|---|---|---|
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI |
InChI=1S/C29H31N7O/c1-21-5-10-25(18-27(21)34-29-31-13-11-26(33-29)24-4-3-12-30-19-24)32-28(37)23-8-6-22(7-9-23)20-36-16-14-35(2)15-17-36/h3-13,18-19H,14-17,20H2,1-2H3,(H,32,37)(H,31,33,34) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI Key |
KTUFNOKKBVMGRW-UHFFFAOYSA-N | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Canonical SMILES |
CC1=C(C=C(C=C1)NC(=O)C2=CC=C(C=C2)CN3CCN(CC3)C)NC4=NC=CC(=N4)C5=CN=CC=C5 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Formula |
C29H31N7O | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
DSSTOX Substance ID |
DTXSID3037125 | |
| Record name | Imatinib | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID3037125 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
Molecular Weight |
493.6 g/mol | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Physical Description |
Solid | |
| Record name | Imatinib | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014757 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Solubility |
Very soluble in water at pH < 5.5 (mesylate salt), 1.46e-02 g/L | |
| Record name | Imatinib | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00619 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Imatinib | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014757 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
CAS No. |
152459-95-5 | |
| Record name | Imatinib | |
| Source | CAS Common Chemistry | |
| URL | https://commonchemistry.cas.org/detail?cas_rn=152459-95-5 | |
| Description | CAS Common Chemistry is an open community resource for accessing chemical information. Nearly 500,000 chemical substances from CAS REGISTRY cover areas of community interest, including common and frequently regulated chemicals, and those relevant to high school and undergraduate chemistry classes. This chemical information, curated by our expert scientists, is provided in alignment with our mission as a division of the American Chemical Society. | |
| Explanation | The data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated. | |
| Record name | Imatinib [INN:BAN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0152459955 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Imatinib | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00619 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Imatinib | |
| Source | DTP/NCI | |
| URL | https://dtp.cancer.gov/dtpstandard/servlet/dwindex?searchtype=NSC&outputformat=html&searchlist=759854 | |
| Description | The NCI Development Therapeutics Program (DTP) provides services and resources to the academic and private-sector research communities worldwide to facilitate the discovery and development of new cancer therapeutic agents. | |
| Explanation | Unless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source. | |
| Record name | Imatinib | |
| Source | DTP/NCI | |
| URL | https://dtp.cancer.gov/dtpstandard/servlet/dwindex?searchtype=NSC&outputformat=html&searchlist=743414 | |
| Description | The NCI Development Therapeutics Program (DTP) provides services and resources to the academic and private-sector research communities worldwide to facilitate the discovery and development of new cancer therapeutic agents. | |
| Explanation | Unless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source. | |
| Record name | Imatinib | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID3037125 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
| Record name | IMATINIB | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/BKJ8M8G5HI | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
| Record name | Imatinib | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014757 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Melting Point |
226 °C (mesylate salt) | |
| Record name | Imatinib | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00619 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Imatinib | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014757 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Synthesis routes and methods I
Procedure details









Synthesis routes and methods II
Procedure details






Synthesis routes and methods III
Procedure details








Synthesis routes and methods IV
Procedure details











Synthesis routes and methods V
Procedure details







Retrosynthesis Analysis
AI-Powered Synthesis Planning: Our tool employs the Template_relevance Pistachio, Template_relevance Bkms_metabolic, Template_relevance Pistachio_ringbreaker, Template_relevance Reaxys, Template_relevance Reaxys_biocatalysis model, leveraging a vast database of chemical reactions to predict feasible synthetic routes.
One-Step Synthesis Focus: Specifically designed for one-step synthesis, it provides concise and direct routes for your target compounds, streamlining the synthesis process.
Accurate Predictions: Utilizing the extensive PISTACHIO, BKMS_METABOLIC, PISTACHIO_RINGBREAKER, REAXYS, REAXYS_BIOCATALYSIS database, our tool offers high-accuracy predictions, reflecting the latest in chemical research and data.
Strategy Settings
| Precursor scoring | Relevance Heuristic |
|---|---|
| Min. plausibility | 0.01 |
| Model | Template_relevance |
| Template Set | Pistachio/Bkms_metabolic/Pistachio_ringbreaker/Reaxys/Reaxys_biocatalysis |
| Top-N result to add to graph | 6 |
Feasible Synthetic Routes
Disclaimer and Information on In-Vitro Research Products
Please be aware that all articles and product information presented on BenchChem are intended solely for informational purposes. The products available for purchase on BenchChem are specifically designed for in-vitro studies, which are conducted outside of living organisms. In-vitro studies, derived from the Latin term "in glass," involve experiments performed in controlled laboratory settings using cells or tissues. It is important to note that these products are not categorized as medicines or drugs, and they have not received approval from the FDA for the prevention, treatment, or cure of any medical condition, ailment, or disease. We must emphasize that any form of bodily introduction of these products into humans or animals is strictly prohibited by law. It is essential to adhere to these guidelines to ensure compliance with legal and ethical standards in research and experimentation.