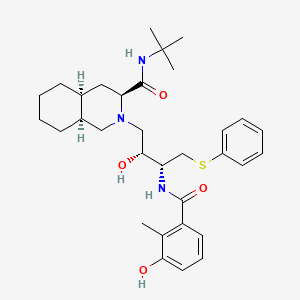
Nelfinavir
Description
This compound is a potent HIV-1 protease inhibitor. It is used in combination with other antiviral drugs in the treatment of HIV in both adults and children. This compound inhibits the HIV viral proteinase enzyme which prevents cleavage of the gag-pol polyprotein, resulting in noninfectious, immature viral particles.
This compound is a Protease Inhibitor. The mechanism of action of this compound is as a HIV Protease Inhibitor, and Cytochrome P450 3A Inhibitor.
This compound is an antiretroviral protease inhibitor used in the therapy and prevention of human immunodeficiency virus (HIV) infection and the acquired immunodeficiency syndrome (AIDS). This compound can cause transient and usually asymptomatic elevations in serum aminotransferase levels and is a rare cause of clinically apparent, acute liver injury. In HBV or HCV coinfected patients, hepatic injury during antiretroviral therapy that includes this compound may be a result of exacerbation of the underlying chronic hepatitis B or C, rather than a direct effect of the medication.
This compound is a synthetic antiviral agent that selectively binds to and inhibits human immunodeficiency virus (HIV) protease. This compound has activity against HIV 1 and 2.
A potent HIV protease inhibitor. It is used in combination with other antiviral drugs in the treatment of HIV in both adults and children.
See also: this compound Mesylate (has salt form).
Properties
IUPAC Name |
(3S,4aS,8aS)-N-tert-butyl-2-[(2R,3R)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylsulfanylbutyl]-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinoline-3-carboxamide | |
|---|---|---|
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI |
InChI=1S/C32H45N3O4S/c1-21-25(15-10-16-28(21)36)30(38)33-26(20-40-24-13-6-5-7-14-24)29(37)19-35-18-23-12-9-8-11-22(23)17-27(35)31(39)34-32(2,3)4/h5-7,10,13-16,22-23,26-27,29,36-37H,8-9,11-12,17-20H2,1-4H3,(H,33,38)(H,34,39)/t22-,23+,26-,27-,29+/m0/s1 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI Key |
QAGYKUNXZHXKMR-HKWSIXNMSA-N | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Canonical SMILES |
CC1=C(C=CC=C1O)C(=O)NC(CSC2=CC=CC=C2)C(CN3CC4CCCCC4CC3C(=O)NC(C)(C)C)O | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Isomeric SMILES |
CC1=C(C=CC=C1O)C(=O)N[C@@H](CSC2=CC=CC=C2)[C@@H](CN3C[C@H]4CCCC[C@H]4C[C@H]3C(=O)NC(C)(C)C)O | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Formula |
C32H45N3O4S | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Related CAS |
159989-65-8 (monomethane sulfonate (salt)) | |
| Record name | Nelfinavir [INN:BAN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0159989647 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
DSSTOX Substance ID |
DTXSID5035080 | |
| Record name | Nelfinavir | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID5035080 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
Molecular Weight |
567.8 g/mol | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Physical Description |
Solid | |
| Record name | Nelfinavir | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014365 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Solubility |
Slightly soluble, 1.91e-03 g/L | |
| Record name | Nelfinavir | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00220 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Nelfinavir | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014365 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
CAS No. |
159989-64-7 | |
| Record name | Nelfinavir | |
| Source | CAS Common Chemistry | |
| URL | https://commonchemistry.cas.org/detail?cas_rn=159989-64-7 | |
| Description | CAS Common Chemistry is an open community resource for accessing chemical information. Nearly 500,000 chemical substances from CAS REGISTRY cover areas of community interest, including common and frequently regulated chemicals, and those relevant to high school and undergraduate chemistry classes. This chemical information, curated by our expert scientists, is provided in alignment with our mission as a division of the American Chemical Society. | |
| Explanation | The data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated. | |
| Record name | Nelfinavir [INN:BAN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0159989647 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Nelfinavir | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00220 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | nelfinavir | |
| Source | DTP/NCI | |
| URL | https://dtp.cancer.gov/dtpstandard/servlet/dwindex?searchtype=NSC&outputformat=html&searchlist=747167 | |
| Description | The NCI Development Therapeutics Program (DTP) provides services and resources to the academic and private-sector research communities worldwide to facilitate the discovery and development of new cancer therapeutic agents. | |
| Explanation | Unless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source. | |
| Record name | Nelfinavir | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID5035080 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
| Record name | NELFINAVIR | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/HO3OGH5D7I | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
| Record name | Nelfinavir | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014365 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Melting Point |
349.84 °C | |
| Record name | Nelfinavir | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00220 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Nelfinavir | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014365 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Advanced Research Methodologies and Future Directions
Omics Technologies in Nelfinavir Research
Omics technologies provide a global view of biological systems, allowing researchers to identify this compound's interactions at the molecular level, including its effects on proteins, genes, and metabolic profiles.
Proteome-wide affinity purification is a powerful technique used to identify direct protein targets and interacting partners of this compound within a cellular context. Studies employing this methodology have revealed that this compound possesses multiple activity-specific binding partners. Many of these partners are embedded within the lipid bilayers of cellular organelles, specifically mitochondria and the endoplasmic reticulum. aacrjournals.orgnih.gov This suggests that this compound's mechanism of action involves direct interaction with membrane-associated proteins.
Furthermore, computational analyses have predicted a broader range of cellular targets for this compound. One such analysis identified 92 putative human off-targets, with the seven strongest binding affinities attributed to aspartyl proteases. nih.govplos.org Beyond proteases, this compound has been shown to directly target human DDI2, an inhibition that effectively blocks NFE2L1 proteolysis and enhances the cytotoxicity of other proteasome inhibitors in cancer cells. biorxiv.org Research also indicates that this compound can inhibit multiple members of the protein kinase-like superfamily, including key signaling molecules such as EGFR, ErbB2, ErbB4, Akt1, Akt2, and Akt3, contributing to its broad anti-cancer effects. plos.org Additionally, this compound has been observed to enhance the interaction between heat shock protein 70 (HSP70) and heat shock protein 90 (HSP90), suggesting HSP90 as another potential molecular target. researchgate.net
Table 1: Key Protein Targets and Interactors of this compound Identified by Proteome-Wide Approaches
| Protein/Protein Family | Associated Cellular Process/Pathway | Reference |
| Membrane-embedded proteins (Mitochondria, ER) | Lipid bilayer stress, mitochondrial respiration, vesicular transport | aacrjournals.orgnih.gov |
| Aspartyl Proteases | Diverse cellular processes, potential off-targets | nih.govplos.org |
| Human DDI2 | NFE2L1 proteolysis, proteasome synthesis | biorxiv.org |
| Protein Kinase-like Superfamily (EGFR, ErbB2, ErbB4, Akt1, Akt2, Akt3) | Cell proliferation, apoptosis, PI3K/Akt pathway, cancer progression | plos.org |
| HSP90 | Protein folding, ER stress, Akt signaling | researchgate.net |
Genome-wide CRISPR/Cas9-based screening is a high-throughput genetic tool used to identify genes that modulate a cell's response to a drug, such as this compound. This methodology, often combined with proteome-wide affinity purification, helps pinpoint genetic contributors that influence this compound's cytotoxicity. aacrjournals.orgnih.govaacrjournals.orgresearchgate.net
A significant finding from such screens is the identification of ADIPOR2 (Adiponectin Receptor 2) as a gene whose sensitivity to this compound is dependent on its function. ADIPOR2 plays a crucial role in maintaining membrane fluidity by promoting fatty acid desaturation and their incorporation into phospholipids. aacrjournals.orgnih.govaacrjournals.org This suggests a direct link between cellular lipid metabolism and this compound's efficacy. CRISPR/Cas9 screens are instrumental in detecting genes essential for cell survival and those conferring drug resistance, offering insights into this compound's mechanism of action and potential resistance pathways. thno.org
Specifically, this compound treatment results in a notable increase in saturated fatty acids (SFA) within membrane phospholipids, including phosphatidylcholines (PC) and phosphatidylethanolamines (PE), while simultaneously decreasing monounsaturated fatty acids (MUFA). nih.gov The compound also modifies the composition of lipid droplets, leading to a decrease in cholesterol esters and an increase in triacylglycerols. nih.gov Global lipidomics analyses have further confirmed these effects, particularly on phosphatidylcholines and phosphatidylinositols, which are critical components of cellular membranes. nih.gov
These metabolic alterations are directly linked to this compound's cellular effects. Supplementation with exogenous fatty acids has been shown to prevent the this compound-induced disruption of mitochondrial metabolism and the activation of stress responses. aacrjournals.orgnih.govnih.gov Conversely, the depletion of fatty acids and cholesterol pools, for instance, by the drug Ezetimibe, demonstrates a synergistic anti-cancer activity with this compound in vitro. aacrjournals.orgnih.govnih.gov These findings collectively highlight that this compound induces lipid bilayer stress in cellular organelles, leading to metabolic rewiring and activation of the unfolded protein response, which are crucial for its broad anti-cancer activity. aacrjournals.orgnih.govnih.gov
Table 2: this compound's Effects on Lipid and Metabolic Profiles
| Lipid/Metabolic Component | Observed Effect | Consequence/Implication | Reference |
| Lipid-rich membranes | Altered fluidity and composition | Disrupted mitochondrial respiration, blocked vesicular transport, lipid bilayer stress | aacrjournals.orgnih.govresearchgate.netnih.gov |
| Saturated Fatty Acids (SFA) in phospholipids | Increased | Altered membrane composition | nih.gov |
| Monounsaturated Fatty Acids (MUFA) in phospholipids | Decreased | Altered membrane composition | nih.gov |
| Cholesterol Esters in lipid droplets | Decreased | Altered lipid droplet composition | nih.gov |
| Triacylglycerols in lipid droplets | Increased | Altered lipid droplet composition | nih.gov |
| Phosphatidylcholines (PC) & Phosphatidylinositols (PI) | Affected composition | Critical for membrane integrity and function | nih.gov |
| Mitochondrial metabolism | Disrupted | Metabolic rewiring, stress response activation | aacrjournals.orgnih.govnih.gov |
Genome-Wide CRISPR/Cas9-Based Screening
In Vitro and In Vivo Model Systems for this compound Studies
To comprehensively evaluate this compound's biological activities, researchers utilize a range of model systems, from isolated cell lines to complex animal models, providing insights into its cellular and systemic effects.
Cell line-based assays are fundamental for initial screening and mechanistic studies of this compound. These assays have extensively demonstrated this compound's anti-cancer properties across a diverse panel of human cancer cell lines, including those derived from lung, breast, prostate, glioblastoma, pediatric leukemia, non-small cell lung cancer (NSCLC), small-cell lung cancer (SCLC), and multiple myeloma. nih.govmdpi.comdovepress.comnih.goviiarjournals.orgiiarjournals.org
Commonly employed assays include cell proliferation assays to measure growth inhibition, cell viability assays (such as the MTT assay) to quantify live cells, and immunofluorescence to visualize cellular components or viral antigens. iiarjournals.orgnih.govmdpi.combiorxiv.orgregenhealthsolutions.infonih.gov Through these methods, this compound has been shown to:
Inhibit cellular proliferation. mdpi.comdovepress.comiiarjournals.org
Induce various forms of cell death, including apoptosis and autophagy. nih.govresearchgate.netmdpi.comdovepress.comnih.goviiarjournals.orgiiarjournals.org
Trigger endoplasmic reticulum (ER) stress and activate the unfolded protein response (UPR). aacrjournals.orgnih.govnih.govresearchgate.netnih.govmdpi.comdovepress.comiiarjournals.orgiiarjournals.org
Interfere with crucial cell signaling pathways, notably the Akt and mTOR pathways. nih.govplos.orgresearchgate.netdovepress.comnih.goviiarjournals.org
Induce cell cycle arrest. mdpi.comdovepress.com
Exhibit cell type-dependent inhibition of proteasome activity. nih.govmdpi.com
Affect the replication of oncogenic herpesviruses, such as Kaposi sarcoma-associated herpesvirus and Epstein-Barr virus. nih.govnih.gov
Beyond cancer, this compound has also demonstrated antiviral activity in cell lines. For instance, it strongly inhibited the replication of SARS-CoV in Vero E6 cells, evidenced by a reduction in the cytopathic effect and decreased viral antigen expression detected by immunofluorescence. nih.govmdpi.combiorxiv.orgregenhealthsolutions.info
Animal models are critical for preclinical evaluation, providing a more complex biological context to assess this compound's efficacy and mechanisms in vivo. This compound has shown significant anti-tumor effects in various mouse xenograft models. researchgate.netmdpi.comiiarjournals.orgiiarjournals.orgresearchgate.net
In human NSCLC xenograft tumors, this compound treatment led to reduced tumor growth, which correlated with increased apoptosis and ER stress within the tumor tissue. iiarjournals.org It also inhibited the growth of SCLC patient-derived xenograft (PDX) mouse models. iiarjournals.org Furthermore, this compound exhibited therapeutic efficacy against T-cell acute lymphoblastic leukemia (T-ALL) in an SCL-LMO1 transgenic mouse model. researchgate.net Combined therapeutic approaches in animal models have also yielded promising results; for example, the combination of this compound and Bortezomib resulted in increased expression of key ER stress markers like CHOP and ATF4 in tumors derived from xenograft models of mTOR hyperactive cells. mdpi.com
Beyond oncology, this compound's potential has been explored in other disease models. In Echinococcus multilocularis PSC-infected BALB/c mice and immunodeficient mice, daily oral administration of this compound significantly reduced parasite burden (cyst weight), demonstrating its efficacy in an infectious disease model. nih.gov
Q & A
Q. What experimental models are commonly used to evaluate nelfinavir's anticancer efficacy, and how do they inform mechanistic insights?
- Methodological Answer : Preclinical studies often use in vitro cancer cell lines (e.g., SCLC, breast cancer) and in vivo patient-derived xenograft (PDX) models. For example:
- Cell viability assays (MTT, flow cytometry) assess apoptosis via Annexin V/PI staining (e.g., MDA-MB231, MCF-7) .
- Western blotting evaluates biomarkers like Bak, cytochrome c, and caspases to confirm apoptosis .
- PDX models validate tumor regression and molecular pathways (e.g., mTOR inhibition, UPR induction) in SCLC .
Q. How is this compound quantified in pharmacokinetic studies, and what analytical methods ensure accuracy?
- Methodological Answer : UV-Visible spectrophotometry (e.g., λ = 210–310 nm) with validation parameters:
- Linearity : 10–60 µg/ml for this compound (R² > 0.999) .
- Calibration : Equations like are derived via least squares regression .
LC-MS/MS is preferred for plasma metabolite quantification (e.g., M8 levels: 0.55–1.96 µM) .
Q. What is the role of this compound's active metabolite M8 in antiviral and anticancer activity?
- Methodological Answer :
- CYP2C19 metabolism : Generates M8, which retains anti-HIV activity (EC₅₀: 34 nM vs. This compound’s 30 nM) and binds PXR competitively .
- Docking simulations : M8 shows distinct binding poses in PXR’s ligand-binding pocket (LBP), influencing receptor antagonism .
Advanced Research Questions
Q. How does this compound modulate oxidative stress to induce cancer cell death, and what experimental approaches validate this?
- Methodological Answer :
- ROS assays : H₂DCF-DA staining quantifies ROS accumulation (e.g., 2–3-fold increase in MDA-MB231 cells) .
- Lipid peroxidation : MDA levels (nM) correlate with this compound-induced oxidative damage .
- Akt pathway disruption : ROS-dependent degradation of Akt-HSP90 complexes, validated via immunoprecipitation .
Q. What computational methods predict this compound’s off-target kinase interactions, and how do they align with experimental data?
- Methodological Answer :
- MM/GBSA free energy calculations : Predict binding affinities for EGFR (ΔG = −50 kcal/mol) but not FGFR/EPHB4 .
- Molecular docking : Identifies ATP-binding site interactions (e.g., EGFR vs. lapatinib) .
Discrepancies arise for Akt2/CDK2, where computational predictions lack experimental validation .
Q. How do pharmacogenomic variants influence this compound’s plasma exposure and clinical outcomes?
- Methodological Answer :
- CYP2B6 516G→T : Correlates with 30% higher efavirenz exposure (p < 0.01) .
- CYP2C19 681G→A : Reduces this compound clearance, linked to 40% lower virologic failure risk (p = 0.039) .
- MDR1 3435 TT genotype : Associated with reduced resistance emergence in HIV trials (OR = 0.6) .
Q. What mechanisms underlie this compound’s radiosensitizing effects, and how are they tested in combination therapies?
- Methodological Answer :
- HIF-1α/VEGF suppression : Western blotting shows this compound reduces HIF-1α by 70% under hypoxia, validated via EF5 hypoxia marker assays .
- In vivo xenografts : Tumor regrowth delay (2–3 weeks) with this compound + radiation vs. radiation alone .
- Synergy with cisplatin : PPP2R1A silencing enhances apoptosis (p < 0.001) in LUAD models .
Methodological Considerations
Q. How are contradictions in this compound’s efficacy across cancer cell lines addressed experimentally?
- Answer :
- Cell line stratification : Compare genetic profiles (e.g., H69 SCLC cells with PIK3CA mutations resist mTOR inhibition) .
- Biomarker panels : Use ATF4, CHOP, and SESN2 to differentiate UPR vs. mTOR-driven responses .
- Pharmacological manipulation : Co-treatment with rapamycin/tunicamycin isolates pathway-specific effects .
Q. What in silico and in vitro assays resolve this compound’s nuclear receptor agonism/antagonism?
- Answer :
- Luciferase reporter assays : HepG2 cells transfected with PXR/CAR show partial agonism (2-fold induction) and competitive antagonism (IC₅₀ = 10 µM) .
- Limited proteolysis : PXR-LBD structural changes confirm ligand binding (30–250 µM this compound) .
- AlphaSphere analysis : Identifies alternative binding sites (e.g., 1M13, 2O9I) via docking simulations .
Retrosynthesis Analysis
AI-Powered Synthesis Planning: Our tool employs the Template_relevance Pistachio, Template_relevance Bkms_metabolic, Template_relevance Pistachio_ringbreaker, Template_relevance Reaxys, Template_relevance Reaxys_biocatalysis model, leveraging a vast database of chemical reactions to predict feasible synthetic routes.
One-Step Synthesis Focus: Specifically designed for one-step synthesis, it provides concise and direct routes for your target compounds, streamlining the synthesis process.
Accurate Predictions: Utilizing the extensive PISTACHIO, BKMS_METABOLIC, PISTACHIO_RINGBREAKER, REAXYS, REAXYS_BIOCATALYSIS database, our tool offers high-accuracy predictions, reflecting the latest in chemical research and data.
Strategy Settings
| Precursor scoring | Relevance Heuristic |
|---|---|
| Min. plausibility | 0.01 |
| Model | Template_relevance |
| Template Set | Pistachio/Bkms_metabolic/Pistachio_ringbreaker/Reaxys/Reaxys_biocatalysis |
| Top-N result to add to graph | 6 |
Feasible Synthetic Routes
Featured Recommendations
| Most viewed | ||
|---|---|---|
| Most popular with customers |
Disclaimer and Information on In-Vitro Research Products
Please be aware that all articles and product information presented on BenchChem are intended solely for informational purposes. The products available for purchase on BenchChem are specifically designed for in-vitro studies, which are conducted outside of living organisms. In-vitro studies, derived from the Latin term "in glass," involve experiments performed in controlled laboratory settings using cells or tissues. It is important to note that these products are not categorized as medicines or drugs, and they have not received approval from the FDA for the prevention, treatment, or cure of any medical condition, ailment, or disease. We must emphasize that any form of bodily introduction of these products into humans or animals is strictly prohibited by law. It is essential to adhere to these guidelines to ensure compliance with legal and ethical standards in research and experimentation.


