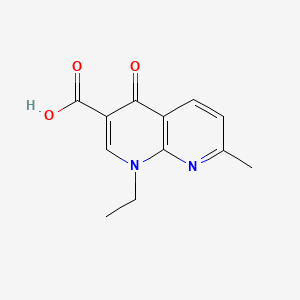
Nalidixic acid
Description
Historical Context of Nalidixic Acid Discovery and Development
The emergence of this compound was a notable event in the search for new antibacterial agents. Its path to discovery was somewhat serendipitous, arising from research initially focused on a different therapeutic area.
Origin as a Byproduct of Chloroquine Synthesis
This compound was discovered in the early 1960s by George Lesher and colleagues at the Sterling-Winthrop Research Institute. patsnap.com This discovery occurred unexpectedly during attempts to synthesize chloroquine, an antimalarial drug. acs.orglums.ac.irnih.govrjptonline.orgringbio.com Specifically, this compound was identified as a byproduct or impurity in the chemical synthesis process of chloroquine. acs.orglums.ac.irnih.govrjptonline.orgringbio.com This "active impurity" served as the lead structure that guided subsequent research efforts, ultimately leading to the identification and development of this compound. acs.orgacs.org
Initial Classification as a Naphthyridone
Chemically, this compound is characterized by a 1,8-naphthyridine nucleus. wikipedia.orgcreative-diagnostics.com This structure contains two nitrogen atoms within its bicyclic ring system, distinguishing it technically as a naphthyridone rather than a quinoline, which possesses a single nitrogen atom in its core structure. wikipedia.orgcreative-diagnostics.com Despite this technical classification, it is widely considered the first of the synthetic quinolone antibiotics due to its structural similarities and mechanism of action shared with the broader class. lums.ac.irwikipedia.orgcreative-diagnostics.com
Early Clinical Introduction for Urinary Tract Infections (UTIs)
Following its discovery and initial characterization, this compound was introduced into clinical practice. wikipedia.orgoup.com Beginning in 1967, it was used for the treatment of bacterial infections. wikipedia.org Its primary early application was in managing uncomplicated urinary tract infections (UTIs). patsnap.comringbio.comwikipedia.orgoup.comoup.comzeelabpharmacy.comtaylorandfrancis.com this compound demonstrated effectiveness particularly against Gram-negative bacteria commonly implicated in UTIs, such as Escherichia coli, Proteus, Shigella, Enterobacter, and Klebsiella species. patsnap.comwikipedia.orgoup.com
Evolution within the Quinolone Class of Antibiotics
This compound's significance extends beyond its initial clinical use; it served as the foundational structure for the development of a substantial class of antibacterial agents.
First-Generation Quinolone
This compound is recognized as the first-generation quinolone antibiotic. wikipedia.orgoup.comnih.govwikipedia.orgmsdvetmanual.comfpnotebook.com Antibiotics within this initial generation, including this compound, were primarily effective against aerobic Gram-negative bacteria. oup.comnih.govwikipedia.org Their spectrum of activity was considered narrow compared to later developments in the class. nih.govedscl.in
Precursor to Fluoroquinolones
The discovery and study of this compound paved the way for the development of subsequent generations of quinolone antibiotics, most notably the fluoroquinolones. acs.orglums.ac.irnih.govwikipedia.orgmsdvetmanual.comedscl.innih.govguidetopharmacology.orgacs.orgslideshare.net Modifications to the basic quinolone structure, such as the addition of a fluorine atom, led to compounds with significantly enhanced antibacterial activity, a broader spectrum encompassing some Gram-positive bacteria, and improved pharmacokinetic properties. oup.comnih.govwikipedia.orgmsdvetmanual.comedscl.in This evolution from this compound to the fluoroquinolones represents a major advancement in antimicrobial therapy. edscl.in
Influence on Subsequent Quinolone Development
The discovery and characterization of this compound were foundational to the development of the entire quinolone class of antibiotics. Despite its limitations, this compound demonstrated the potential of targeting bacterial DNA replication machinery as a therapeutic strategy. The challenges encountered with this compound, particularly its narrow spectrum and the emergence of resistance, spurred extensive research efforts to synthesize analogs with improved properties nih.gov.
Since the introduction of this compound, over 10,000 analogs have been synthesized, although only a small number have reached clinical practice wikipedia.orgnih.gov. The key breakthrough in the development of more potent and broad-spectrum quinolones came with structural modifications, most notably the addition of a fluorine atom at the C-6 position of the bicyclic core structure, leading to the designation of "fluoroquinolones" wikipedia.orgmicrobiologyresearch.orgrjptonline.orgasm.org. This modification, first seen in compounds like norfloxacin (patented in 1978), significantly expanded the antibacterial spectrum to include many Gram-positive bacteria and improved pharmacokinetic properties, such as better oral absorption and tissue distribution wikipedia.orgnih.govnih.govrjptonline.orgnih.gov.
Further modifications at other positions of the quinolone structure, such as the C-7 and C-8 positions and the N-1 substituent, led to subsequent generations of quinolones with enhanced activity against specific pathogens, improved potency against topoisomerase IV, and altered pharmacokinetic profiles microbiologyresearch.orgasm.orgoup.com. For instance, modifications at the C-7 position have been shown to influence antibacterial potency, spectrum, and even safety profiles asm.orgresearchgate.net. The development trajectory moved from first-generation agents like this compound, oxolinic acid, pipemidic acid, and cinoxacin, which were primarily limited to urinary tract infections caused by Gram-negative bacteria, to second, third, and fourth-generation fluoroquinolones with increasingly broader spectra and improved systemic activity wikipedia.orgnih.govoup.com.
The detailed research into how this compound inhibited DNA gyrase provided a crucial target for rational drug design. Subsequent research focused on developing compounds with increased binding affinity to bacterial topoisomerases and better cellular penetration, overcoming some of the limitations of the parent compound asm.org. The understanding of resistance mechanisms to this compound, particularly mutations in gyrA, also informed the design of newer quinolones aimed at mitigating the development of resistance patsnap.com.
The evolution of quinolone antibiotics from this compound can be illustrated by the development timeline and the changes in their antibacterial spectrum and pharmacokinetic properties across generations.
Table 1: Evolution of Quinolone Generations (Illustrative Data)
| Generation | Key Compounds (Examples) | Primary Spectrum | Key Structural Modifications (relative to this compound) |
| First Generation | This compound, Oxolinic Acid, Cinoxacin | Primarily Gram-negative (UTIs) | Basic naphthyridone/quinolone structure |
| Second Generation | Norfloxacin, Ciprofloxacin, Ofloxacin | Expanded Gram-negative, some Gram-positive | Fluorine at C-6, Piperazine or other group at C-7 |
| Third Generation | Levofloxacin, Gatifloxacin, Moxifloxacin | Improved Gram-positive, broader spectrum | Further modifications at C-7, C-8, or N-1 |
| Fourth Generation | Trovafloxacin (Note: withdrawn/restricted) | Broadest spectrum, including anaerobes | Additional modifications |
This table is illustrative and based on the general characteristics of each generation as described in the literature. An interactive version could allow users to filter by generation or compound.
The research findings on this compound's interaction with bacterial DNA gyrase, its spectrum of activity, and the mechanisms of resistance were fundamental in guiding the synthesis and evaluation of thousands of subsequent quinolone derivatives. The progression from a compound primarily effective against uncomplicated UTIs to potent, broad-spectrum systemic antibacterial agents is a direct consequence of the scientific understanding gained from studying this compound.
Properties
IUPAC Name |
1-ethyl-7-methyl-4-oxo-1,8-naphthyridine-3-carboxylic acid | |
|---|---|---|
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI |
InChI=1S/C12H12N2O3/c1-3-14-6-9(12(16)17)10(15)8-5-4-7(2)13-11(8)14/h4-6H,3H2,1-2H3,(H,16,17) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI Key |
MHWLWQUZZRMNGJ-UHFFFAOYSA-N | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Canonical SMILES |
CCN1C=C(C(=O)C2=C1N=C(C=C2)C)C(=O)O | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Formula |
C12H12N2O3 | |
| Record name | NALIDIXIC ACID | |
| Source | CAMEO Chemicals | |
| URL | https://cameochemicals.noaa.gov/chemical/20720 | |
| Description | CAMEO Chemicals is a chemical database designed for people who are involved in hazardous material incident response and planning. CAMEO Chemicals contains a library with thousands of datasheets containing response-related information and recommendations for hazardous materials that are commonly transported, used, or stored in the United States. CAMEO Chemicals was developed by the National Oceanic and Atmospheric Administration's Office of Response and Restoration in partnership with the Environmental Protection Agency's Office of Emergency Management. | |
| Explanation | CAMEO Chemicals and all other CAMEO products are available at no charge to those organizations and individuals (recipients) responsible for the safe handling of chemicals. However, some of the chemical data itself is subject to the copyright restrictions of the companies or organizations that provided the data. | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Related CAS |
3374-05-8 (hydrochloride salt, anhydrous) | |
| Record name | Nalidixic acid [USAN:USP:INN:BAN:JAN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0000389082 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
DSSTOX Substance ID |
DTXSID3020912 | |
| Record name | Nalidixic acid | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID3020912 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
Molecular Weight |
232.23 g/mol | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Physical Description |
Nalidixic acid is a cream-colored powder. (NTP, 1992), Solid | |
| Record name | NALIDIXIC ACID | |
| Source | CAMEO Chemicals | |
| URL | https://cameochemicals.noaa.gov/chemical/20720 | |
| Description | CAMEO Chemicals is a chemical database designed for people who are involved in hazardous material incident response and planning. CAMEO Chemicals contains a library with thousands of datasheets containing response-related information and recommendations for hazardous materials that are commonly transported, used, or stored in the United States. CAMEO Chemicals was developed by the National Oceanic and Atmospheric Administration's Office of Response and Restoration in partnership with the Environmental Protection Agency's Office of Emergency Management. | |
| Explanation | CAMEO Chemicals and all other CAMEO products are available at no charge to those organizations and individuals (recipients) responsible for the safe handling of chemicals. However, some of the chemical data itself is subject to the copyright restrictions of the companies or organizations that provided the data. | |
| Record name | Nalidixic Acid | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014917 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Solubility |
less than 1 mg/mL at 70 °F (NTP, 1992), Soly at 23 °C (mg/ml): chloroform 35; toluene 1.6; methanol 1.3; ethanol 0.6; water 0.1; ether 0.1., PRACTICALLY INSOL IN WATER; SOL IN SOLN OF CARBONATES, 2.30e+00 g/L | |
| Record name | NALIDIXIC ACID | |
| Source | CAMEO Chemicals | |
| URL | https://cameochemicals.noaa.gov/chemical/20720 | |
| Description | CAMEO Chemicals is a chemical database designed for people who are involved in hazardous material incident response and planning. CAMEO Chemicals contains a library with thousands of datasheets containing response-related information and recommendations for hazardous materials that are commonly transported, used, or stored in the United States. CAMEO Chemicals was developed by the National Oceanic and Atmospheric Administration's Office of Response and Restoration in partnership with the Environmental Protection Agency's Office of Emergency Management. | |
| Explanation | CAMEO Chemicals and all other CAMEO products are available at no charge to those organizations and individuals (recipients) responsible for the safe handling of chemicals. However, some of the chemical data itself is subject to the copyright restrictions of the companies or organizations that provided the data. | |
| Record name | Nalidixic acid | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00779 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | NALIDIXIC ACID | |
| Source | Hazardous Substances Data Bank (HSDB) | |
| URL | https://pubchem.ncbi.nlm.nih.gov/source/hsdb/3241 | |
| Description | The Hazardous Substances Data Bank (HSDB) is a toxicology database that focuses on the toxicology of potentially hazardous chemicals. It provides information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. The information in HSDB has been assessed by a Scientific Review Panel. | |
| Record name | Nalidixic Acid | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014917 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Color/Form |
PALE BUFF, CRYSTALLINE POWDER, WHITE TO SLIGHTLY YELLOW, CRYSTALLINE POWDER | |
CAS No. |
389-08-2 | |
| Record name | NALIDIXIC ACID | |
| Source | CAMEO Chemicals | |
| URL | https://cameochemicals.noaa.gov/chemical/20720 | |
| Description | CAMEO Chemicals is a chemical database designed for people who are involved in hazardous material incident response and planning. CAMEO Chemicals contains a library with thousands of datasheets containing response-related information and recommendations for hazardous materials that are commonly transported, used, or stored in the United States. CAMEO Chemicals was developed by the National Oceanic and Atmospheric Administration's Office of Response and Restoration in partnership with the Environmental Protection Agency's Office of Emergency Management. | |
| Explanation | CAMEO Chemicals and all other CAMEO products are available at no charge to those organizations and individuals (recipients) responsible for the safe handling of chemicals. However, some of the chemical data itself is subject to the copyright restrictions of the companies or organizations that provided the data. | |
| Record name | Nalidixic acid | |
| Source | CAS Common Chemistry | |
| URL | https://commonchemistry.cas.org/detail?cas_rn=389-08-2 | |
| Description | CAS Common Chemistry is an open community resource for accessing chemical information. Nearly 500,000 chemical substances from CAS REGISTRY cover areas of community interest, including common and frequently regulated chemicals, and those relevant to high school and undergraduate chemistry classes. This chemical information, curated by our expert scientists, is provided in alignment with our mission as a division of the American Chemical Society. | |
| Explanation | The data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated. | |
| Record name | Nalidixic acid [USAN:USP:INN:BAN:JAN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0000389082 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Nalidixic acid | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00779 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | nalidixic acid | |
| Source | DTP/NCI | |
| URL | https://dtp.cancer.gov/dtpstandard/servlet/dwindex?searchtype=NSC&outputformat=html&searchlist=757432 | |
| Description | The NCI Development Therapeutics Program (DTP) provides services and resources to the academic and private-sector research communities worldwide to facilitate the discovery and development of new cancer therapeutic agents. | |
| Explanation | Unless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source. | |
| Record name | nalidixic acid | |
| Source | DTP/NCI | |
| URL | https://dtp.cancer.gov/dtpstandard/servlet/dwindex?searchtype=NSC&outputformat=html&searchlist=82174 | |
| Description | The NCI Development Therapeutics Program (DTP) provides services and resources to the academic and private-sector research communities worldwide to facilitate the discovery and development of new cancer therapeutic agents. | |
| Explanation | Unless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source. | |
| Record name | Nalidixic acid | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID3020912 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
| Record name | Nalidixic acid | |
| Source | European Chemicals Agency (ECHA) | |
| URL | https://echa.europa.eu/substance-information/-/substanceinfo/100.006.241 | |
| Description | The European Chemicals Agency (ECHA) is an agency of the European Union which is the driving force among regulatory authorities in implementing the EU's groundbreaking chemicals legislation for the benefit of human health and the environment as well as for innovation and competitiveness. | |
| Explanation | Use of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page. | |
| Record name | NALIDIXIC ACID | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/3B91HWA56M | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
| Record name | NALIDIXIC ACID | |
| Source | Hazardous Substances Data Bank (HSDB) | |
| URL | https://pubchem.ncbi.nlm.nih.gov/source/hsdb/3241 | |
| Description | The Hazardous Substances Data Bank (HSDB) is a toxicology database that focuses on the toxicology of potentially hazardous chemicals. It provides information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. The information in HSDB has been assessed by a Scientific Review Panel. | |
| Record name | Nalidixic Acid | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014917 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Melting Point |
444 to 446 °F (NTP, 1992), 229-230 °C, 229.5 °C | |
| Record name | NALIDIXIC ACID | |
| Source | CAMEO Chemicals | |
| URL | https://cameochemicals.noaa.gov/chemical/20720 | |
| Description | CAMEO Chemicals is a chemical database designed for people who are involved in hazardous material incident response and planning. CAMEO Chemicals contains a library with thousands of datasheets containing response-related information and recommendations for hazardous materials that are commonly transported, used, or stored in the United States. CAMEO Chemicals was developed by the National Oceanic and Atmospheric Administration's Office of Response and Restoration in partnership with the Environmental Protection Agency's Office of Emergency Management. | |
| Explanation | CAMEO Chemicals and all other CAMEO products are available at no charge to those organizations and individuals (recipients) responsible for the safe handling of chemicals. However, some of the chemical data itself is subject to the copyright restrictions of the companies or organizations that provided the data. | |
| Record name | Nalidixic acid | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00779 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | NALIDIXIC ACID | |
| Source | Hazardous Substances Data Bank (HSDB) | |
| URL | https://pubchem.ncbi.nlm.nih.gov/source/hsdb/3241 | |
| Description | The Hazardous Substances Data Bank (HSDB) is a toxicology database that focuses on the toxicology of potentially hazardous chemicals. It provides information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. The information in HSDB has been assessed by a Scientific Review Panel. | |
| Record name | Nalidixic Acid | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014917 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Structure-activity Relationships Sar and Derivative Development
Core Structural Features of Nalidixic Acid
This compound possesses a core bicyclic structure that is fundamental to its antibacterial activity. nih.govnih.govnih.govnih.gov
1,8-Naphthyridine Nucleus
Technically classified as a naphthyridone rather than a quinolone, this compound is characterized by a 1,8-naphthyridine nucleus. wikipedia.orgcreative-diagnostics.comdiscofinechem.comhimedialabs.com This bicyclic system contains two nitrogen atoms within the fused rings, distinguishing it from the quinoline nucleus which has a single nitrogen atom. wikipedia.orgcreative-diagnostics.comdiscofinechem.comhimedialabs.com The core structure includes a carboxylic acid group at position 3 and a keto group at position 4. researchgate.net These functional groups are essential for the interaction with bacterial DNA gyrase and topoisomerase IV, the primary targets of quinolone antibiotics. wikipedia.orgnih.govwikipedia.orgcreative-diagnostics.comdiscofinechem.com this compound itself has an ethyl group at the N-1 position and a methyl group at the C-7 position. researchgate.net
Modifications Leading to Improved Quinolones and Fluoroquinolones
The limited activity and pharmacokinetic properties of this compound spurred extensive research into structural modifications to improve its antibacterial profile. nih.govrjptonline.orgnih.govmicrobiologyresearch.orgbrieflands.com Over 10,000 analogues of this compound and fluoroquinolones have been synthesized. nih.govmicrobiologyresearch.orgbrieflands.com Modifications at key positions on the naphthyridine or quinoline core have led to compounds with enhanced potency, broader spectrum of activity, and improved pharmacokinetic properties. rjptonline.orgnih.govmicrobiologyresearch.orgasm.org The core structure amenable to modification includes a 1-substituted 1,4-dihydro-4-oxo-pyridine-3-carboxylic acid moiety combined with a second aromatic or heteroaromatic ring. asm.org
Substitutions at N-1 Position
The substituent at the N-1 position of the core structure significantly influences antibacterial activity and pharmacokinetic properties. In this compound, this position is occupied by an ethyl group. researchgate.net Modifications at this position have explored various substituents. For instance, a cyclopropyl group at N-1, as seen in ciprofloxacin, confers significant activity, particularly against Gram-negative bacteria. microbiologyresearch.orgasm.org Other groups like 2,4-difluorophenyl have also been investigated. microbiologyresearch.org
Introduction of Fluorine at C-6 Position (Fluoroquinolones)
A pivotal breakthrough in quinolone development was the introduction of a fluorine atom at the C-6 position of the bicyclic ring system. rjptonline.orgnih.govmicrobiologyresearch.orgasm.orgnih.govnih.govresearchgate.netnih.govnih.govaujmsr.com This modification led to the class of fluoroquinolones, which exhibit a significantly broader spectrum of antibacterial activity, including improved activity against Gram-positive bacteria, compared to the earlier quinolones like this compound. nih.govrjptonline.orgwikipedia.orgnih.govmicrobiologyresearch.org The fluorine atom at C-6 enhances the compound's potency by increasing its lipophilicity and improving its interaction with the bacterial DNA gyrase and topoisomerase IV enzymes. asm.org
Substituents at C-7 Position
Modifications at the C-7 position have a profound impact on the antibacterial spectrum and potency, as well as pharmacokinetic properties. asm.orgnih.gov In this compound, a methyl group is present at C-7. researchgate.net The introduction of a piperazine ring at the C-7 position, as found in ciprofloxacin and norfloxacin, is a common modification that significantly enhances activity against Gram-negative bacteria, including Pseudomonas aeruginosa. rjptonline.orgmicrobiologyresearch.orgasm.orgnih.govnih.govnih.govfishersci.cafishersci.ca197.255.126uni.lumims.com Various other nitrogen-containing heterocycles have also been explored at this position, contributing to variations in spectrum and potency. asm.orgaujmsr.com For example, moxifloxacin features a diazabicyclo[3.1.0]hexan-6-yl moiety at C-7, contributing to its expanded spectrum including anaerobic coverage. nih.govnih.govuni.luguidetopharmacology.org
Modifications at C-8 Position
Modifications at the C-8 position can influence the spectrum of activity, phototoxicity, and pharmacokinetic properties. asm.orgguidetopharmacology.org In some later generation fluoroquinolones, a methoxy group or a fluorine atom has been introduced at C-8. For instance, moxifloxacin has a methoxy group at C-8, which contributes to its improved activity against Gram-positive bacteria and atypical pathogens. nih.govnih.govuni.luguidetopharmacology.org The presence of a fluorine at C-8, as seen in some fluoroquinolones like garenoxacin (though garenoxacin has a difluoromethoxy at position 8), can influence the activity profile. brieflands.com
The cumulative effect of these structural modifications, particularly the introduction of fluorine at C-6 and the variation of substituents at N-1 and C-7, has resulted in the diverse range of quinolone and fluoroquinolone antibiotics with varying antibacterial spectra and pharmacological profiles available today.
| Compound Name | PubChem CID | Key Structural Features Relative to this compound |
| This compound | 4421 | 1,8-naphthyridine nucleus, Ethyl at N-1, Methyl at C-7, Carboxylic acid at C-3, Keto at C-4 wikipedia.orgnih.govresearchgate.net |
| Ciprofloxacin | 2764 | Fluoro at C-6, Piperazinyl at C-7, Cyclopropyl at N-1, Quinolone nucleus asm.orgnih.govfishersci.ca197.255.126 |
| Norfloxacin | 4539 | Fluoro at C-6, Piperazinyl at C-7, Ethyl at N-1, Quinolone nucleus asm.orgnih.govnih.govfishersci.cauni.lu |
| Ofloxacin | 4583 | Fluoro at C-6, N-methylpiperazinyl incorporated into a tricyclic ring at C-7, Methyl at C-3 (part of tricyclic), Racemate nih.govnih.govmims.coms4science.atfishersci.ca |
| Levofloxacin | 149096 | (S)-enantiomer of Ofloxacin nih.govnih.govwikidata.orgfishersci.cauni.lu |
| Moxifloxacin | 152946 | Fluoro at C-6, Diazabicyclo[3.1.0]hexan-6-yl at C-7, Methoxy at C-8, Cyclopropyl at N-1, Quinolone nucleus nih.govuni.luguidetopharmacology.org |
| This compound sodium salt | 3864541 | Sodium salt of this compound citeab.com |
| Ciprofloxacin HCl | 62999 | Hydrochloride salt of Ciprofloxacin nih.govciteab.com |
| Moxifloxacin Hydrochloride | 101526 | Hydrochloride salt of Moxifloxacin nih.gov |
| This compound Methyl Ester | 11220033 | Methyl ester at the C-3 carboxylic acid of this compound ontosight.ai |
Impact of Structural Changes on Antimicrobial Activity Spectrum and Potency
The antimicrobial activity of this compound and its derivatives is intrinsically linked to their chemical structure. This compound, as the first-generation quinolone (though technically a naphthyridone), established a foundational structure for subsequent antibiotic development. Its core structure, a 1,4-dihydro-4-oxo-pyridin-3-carboxylic acid moiety annulated with an aromatic ring (specifically a 1,8-naphthyridine nucleus in this compound), is essential for antibacterial activity. slideshare.netaujmsr.combrieflands.com The 3-carboxyl group and the 4-oxo group are particularly crucial, forming a pharmacophore that interacts with bacterial DNA gyrase and topoisomerase IV, the primary targets of this drug class. aujmsr.commdpi.comwikipedia.org
Structural modifications at various positions of the quinolone nucleus have profoundly impacted the antimicrobial spectrum and potency of derivatives compared to the parent compound, this compound. aujmsr.comnih.govnih.govnih.govqeios.com this compound itself exhibits a narrow spectrum, primarily effective against Gram-negative bacteria, including E. coli, Enterobacter, Klebsiella, and Proteus species, and shows minimal activity against Gram-positive organisms. wikipedia.orgmdpi.comnih.gov Its use was largely limited to uncomplicated urinary tract infections due to low serum levels and rapid resistance development. nih.govmdpi.comaafp.org
Key structural changes and their effects on activity include:
Substituent at N-1 Position: The nature of the substituent at the N-1 position significantly influences activity. Optimal substituents often include ethyl, cyclopropyl, and difluorophenyl groups, leading to more potent compounds. nih.govpharmacy180.com For instance, the introduction of a fluorine atom into the N-1 cyclopropyl or 1-butyl substituent has been shown to improve activity against Gram-positive bacteria. pharmacy180.com While steric bulk was initially considered a primary factor, further research indicates a more complex role for N-1 substituents in the mechanism of action. nih.gov
Substitutions at C-2: Simple replacement of the hydrogen at C-2 has generally been detrimental to antibacterial activity. pharmacy180.com
Modifications of the C-3 Carboxylic Acid Group: The carboxylic acid group at C-3 is vital for activity. Modifications typically lead to a decrease in antibacterial potency. pharmacy180.com However, replacing it with an isothiazolo group has resulted in highly active isothiazolo quinolones, demonstrating significantly greater in vitro antibacterial activity than some later-generation quinolones like ciprofloxacin. pharmacy180.com
The C-4-oxo Group: The presence of the 4-oxo group is essential for antibacterial activity. Replacing it with a thioxo or sulfonyl group results in a loss of activity. pharmacy180.com
Substitutions at C-5 Position: Incorporating a group at the C-5 position can be beneficial for antibacterial activity. Studies suggest an order of activity based on the substituent: NH₂ > CH₃ > F, H > OH, or SH, SR. pharmacy180.com
Substitutions at C-6 Position: The introduction of a fluorine atom at the C-6 position was a monumental development, leading to the fluoroquinolone class. pharmacy180.comnih.gov Fluorination at C-6 significantly enhances antibacterial activity and improves penetrability through bacterial membranes in both Gram-positive and Gram-negative bacteria. mdpi.compharmacy180.com This modification broadened the spectrum considerably compared to this compound. nih.gov
Substitutions at C-7 Position: The C-7 position is a key site for structural modifications and has a major impact on activity, spectrum, and pharmacokinetic properties. mdpi.comnih.gov The introduction of a piperazine moiety at C-7 is often essential for activity, and other aminopyrrolidine rings are also compatible. pharmacy180.com Bulky molecules and heterocycles like piperazine, pyrrolidinyl, pyrimidine, indole, or imidazole at this position can modify and often enhance antimicrobial activity, including increasing activity against Gram-positive bacteria. mdpi.comnih.govpharmacy180.com A carbon-nitrogen bond (C7-N) between the nucleus and the substituent is recommended to maintain the antibiotic effect. mdpi.com
Substitutions at C-8 Position: Substituents at C-8 also influence activity. A fluorine substituent at C-8 generally offers good potency against Gram-negative pathogens, while a methoxy group at C-8 can enhance activity against Gram-positive bacteria. pharmacy180.com
The cumulative effect of these structural modifications is evident in the development of subsequent quinolone generations. While this compound (first generation) had limited Gram-negative coverage, second-generation quinolones (e.g., norfloxacin, ciprofloxacin) with a fluorine at C-6 and often a piperazine at C-7 showed increased Gram-negative activity and gained some Gram-positive coverage. slideshare.netaafp.orgnih.govslideshare.net Further modifications in later generations (third and fourth) continued to broaden the spectrum, enhancing activity against Gram-positive organisms, atypical bacteria, and anaerobes. nih.govaafp.orgslideshare.net
Detailed research findings often involve synthesizing novel derivatives and evaluating their Minimum Inhibitory Concentration (MIC) values against a panel of bacteria to quantify the impact of structural changes on potency. For example, studies on novel this compound-based 1,2,4-triazole derivatives have shown that many exhibit better antimicrobial activity than the parent compound, with specific derivatives showing promising activity (e.g., a compound with an MIC of 16 μg/mL). nih.gov Another study involving hydrazine substituted tricyclic derivatives of this compound revealed potent antibacterial activity, particularly against Gram-positive bacteria, with potentiation also observed against Gram-negative bacteria. rroij.com
While specific comprehensive data tables detailing the MICs of a wide range of this compound derivatives with various structural modifications were not extensively detailed across the provided search results, the general principles of SAR are clearly outlined. The impact of key substitutions at positions N-1, C-6, C-7, and C-8 on expanding the spectrum from primarily Gram-negative (this compound) to broader coverage including Gram-positive and other pathogens in later generations is a consistent finding. slideshare.netnih.govaafp.orgnih.govslideshare.net
Below is a conceptual representation of how structural changes influence activity, based on the discussed SAR principles:
| Position | Typical Substituent in this compound | Common Modifications in Derivatives | Impact on Activity/Spectrum |
| N-1 | Ethyl | Cyclopropyl, Fluoroethyl, Methylamino, Methoxy, tert-Butyl, Phenyl | Influence potency; cyclopropyl and fluoroethyl can enhance Gram-positive activity. nih.govpharmacy180.com |
| C-3 | Carboxylic acid | Isothiazolo group | Essential for activity; isothiazolo can significantly increase potency. pharmacy180.com |
| C-4 | Oxo | Thioxo, Sulfonyl | Essential for activity; replacement leads to loss of activity. pharmacy180.com |
| C-6 | Hydrogen | Fluorine | Monumental increase in potency and spectrum; improves penetration. mdpi.compharmacy180.comnih.gov |
| C-7 | Methyl | Piperazine, Aminopyrrolidines, other heterocycles | Major impact on potency and spectrum; crucial for broader Gram-negative and Gram-positive coverage. mdpi.comnih.govpharmacy180.com |
| C-8 | Hydrogen | Fluorine, Methoxy, Chlorine | C-8 F: good Gram-negative potency; C-8 OCH₃: active against Gram-positive. pharmacy180.com |
This table summarizes the general trends observed in the SAR of this compound and its derivatives, highlighting how specific modifications contribute to enhanced or altered antimicrobial profiles. The development of subsequent quinolone generations is a direct result of systematic structural modifications aimed at improving potency, broadening the spectrum, and enhancing pharmacokinetic properties compared to the foundational compound, this compound. aujmsr.comnih.govnih.gov
Interactions with Other Agents and in Vitro Studies
Drug-Drug Interactions and Their Mechanisms
The coadministration of nalidixic acid with certain drugs can lead to altered absorption, metabolism, or excretion, resulting in modified systemic or urinary concentrations of the antibiotic.
Coadministration with Probenecid
Probenecid, a uricosuric agent, is known to interact with organic acids, including certain antibiotics. This interaction has notable effects on the renal handling of this compound.
Probenecid inhibits the tubular secretion of this compound in the kidneys. fda.govnih.gov This occurs through competitive inhibition of the renal organic anion transporter system, which is responsible for the active secretion of organic acids from the blood into the renal tubules. drugs.comnih.gov A pilot study demonstrated that under probenecid co-medication, the renal clearance of both this compound and its metabolite, 7-hydroxymethylthis compound, were reduced. nih.gov
The inhibition of renal tubular secretion by probenecid leads to decreased excretion of this compound in the urine. fda.govdrugs.com This can result in reduced concentrations of the active drug within the urinary tract, potentially lowering its efficacy in treating urinary tract infections. fda.gov Conversely, the decreased renal clearance can lead to increased and prolonged systemic plasma concentrations of this compound. fda.govdrugbank.com
A pilot study showed that under probenecid co-medication, the renal glucuronidation of this compound was reduced from 53% to 16%. nih.gov The intrinsic half-life of the metabolite 7-hydroxymethylthis compound also increased significantly from 0.48 hours to 4.24 hours. nih.gov
Inhibition of Renal Tubular Secretion of this compound
Interactions with Cations (Magnesium, Aluminum, Calcium, Iron, Zinc)
This compound interacts with various multivalent metal cations, which can impact its absorption from the gastrointestinal tract.
Oral preparations containing multivalent cations such as magnesium, aluminum, or calcium can significantly decrease the gastrointestinal absorption of this compound. drugs.comaafp.orgdrugs.comdrugs.commedindia.net This reduction in absorption is due to the formation of stable chelation complexes between this compound and the polyvalent cations in the gastrointestinal tract. drugs.comaafp.org These complexes are typically poorly soluble and not readily absorbed, leading to lower systemic drug concentrations. drugs.comaafp.org Interactions have also been reported with other polyvalent cations like iron and zinc. drugs.comaafp.org Studies have shown that quinolones, including this compound, chelate with these cations, and this interaction significantly reduces absorption and bioavailability, resulting in lower serum drug concentrations. aafp.orgnih.gov The stoichiometry of the this compound-aluminum ion complex has been estimated to be 3 to 1. jst.go.jp
Effects on Theophylline and Caffeine Metabolism
This compound has been shown to interfere with the metabolism of certain methylxanthines, specifically theophylline and caffeine.
Quinolones, including this compound, can increase the plasma concentrations and pharmacologic effects of caffeine due to the inhibition of the CYP450 1A2 enzyme, which is involved in caffeine metabolism. drugs.comdrugs.comrxlist.comdrugs.com This can lead to reduced clearance of caffeine and a prolongation of its plasma half-life. rxlist.com Similarly, the metabolism of theophylline can be decreased when combined with this compound, potentially leading to elevated plasma levels of theophylline. fda.govrxlist.comdrugbank.comnih.gov
The interaction with theophylline has been linked to the structural features of certain quinolones. Quinolones with a naphthyridine ring, like this compound and enoxacin, appear more likely to interact with theophylline compared to those lacking a nitrogen at position 8, such as ciprofloxacin, pefloxacin, and ofloxacin. asm.orgoup.com This suggests that the naphthyridine ring may be an important structural component influencing the inhibition of theophylline metabolism. asm.org The nitrogen atom in the 7-piperazinyl group, if present, is also considered essential for the interaction with cytochrome P-450, the enzyme system that metabolizes theophylline. oup.com
Enhancement of Oral Anticoagulant Effects (e.g., Warfarin)
Concurrent administration of this compound with oral anticoagulants, such as warfarin, has been reported to enhance the anticoagulant effect patsnap.comnih.govpediatriconcall.comrxlist.commims.com. This interaction can lead to an increased risk of bleeding patsnap.comproquest.com. Several potential mechanisms have been proposed for this enhancement:
Displacement from Plasma Proteins: this compound may displace warfarin from its plasma protein binding sites, increasing the concentration of free, active warfarin in the bloodstream nih.govacpjournals.orgjst.go.jp. Warfarin is highly bound to plasma proteins, and even small changes in protein binding can significantly alter its anticoagulant effect drugbank.comjst.go.jpuneb.br.
Influence on Vitamin K-Producing Bacteria: Fluoroquinolones, including the class to which this compound belongs, may affect the gut bacteria that produce vitamin K. nih.gov. Vitamin K is essential for the synthesis of clotting factors, and a reduction in its availability can potentiate the effects of warfarin, which acts by inhibiting vitamin K epoxide reductase uneb.br.
Inhibition of Warfarin Metabolism: While some fluoroquinolones are known to inhibit hepatic cytochrome P450 enzymes responsible for warfarin metabolism, leading to reduced clearance and increased plasma levels of warfarin, the specific effect of this compound on these enzymes, particularly CYP1A2, has also been noted nih.govuneb.br.
Case reports have documented significant increases in the International Normalized Ratio (INR), a measure of blood clotting time, following the initiation of this compound therapy in patients stabilized on warfarin nih.govacpjournals.org. For instance, one case reported an increase in INR from 1.9 to 9.6 in an elderly patient after starting this compound nih.gov. Another case involved gastrointestinal haemorrhage in an elderly patient receiving concomitant warfarin and this compound proquest.com. The susceptibility to this interaction may be influenced by factors such as age, sex, or concomitant diseases nih.gov.
Antimicrobial Synergy and Antagonism
The interaction between this compound and other antimicrobial agents in vitro can result in synergistic (enhanced activity) or antagonistic (reduced activity) effects, depending on the specific combination of drugs and the bacterial strain being tested.
Antagonism with Bacteriostatic Agents (e.g., Tetracycline, Chloramphenicol, Nitrofurantoin)
In vitro studies have consistently demonstrated that bacteriostatic agents, such as tetracycline, chloramphenicol, and nitrofurantoin, can antagonize the bactericidal action of this compound rxlist.comoup.comoup.comrwandafda.gov.rwpharmacy180.comcabidigitallibrary.orgptbioch.edu.plmicrobiologyresearch.org. This compound's bactericidal activity is dependent on active bacterial growth and protein synthesis oup.commicrobiologyresearch.org. Bacteriostatic antibiotics inhibit bacterial growth and protein synthesis, thereby interfering with the conditions necessary for this compound to exert its full killing effect oup.commicrobiologyresearch.orgresearchgate.net.
Studies using techniques like plate diffusion tests and viable counts have shown that the presence of bacteriostatic concentrations of these agents can reduce or abolish the bactericidal effect of this compound against various Gram-negative bacilli, including Escherichia coli and Proteus mirabilis oup.comoup.commicrobiologyresearch.orgnih.gov. For example, chloramphenicol has been shown to totally inhibit the bactericidal activity of this compound against E. coli in nutrient broth microbiologyresearch.org. Similarly, nitrofurantoin has been shown to antagonize the activity of this compound against many Gram-negative bacilli oup.comrwandafda.gov.rwpharmacy180.comnih.gov.
The antagonism between this compound and nitrofurantoin has been known for many years and demonstrated in vitro against certain Gram-negative bacilli oup.com. This antagonism is thought to be associated with the abolition of the bacteriolytic response to quinolones oup.com.
Synergistic Interactions with Tetracycline in Resistant Strains
While antagonism is typically observed with bacteriostatic agents, research has also revealed instances of synergistic interactions between this compound and tetracycline, particularly against multidrug-resistant (MDR) bacterial strains nih.govnih.govresearchgate.netresearchgate.netnih.gov. This synergistic effect appears to be strain-specific and can differ significantly between susceptible and resistant isolates nih.govnih.govresearchgate.net.
Studies using checkerboard assays and time-kill curve analysis have shown that the combination of this compound and tetracycline can exhibit synergy against clinical MDR isolates of Acinetobacter baumannii and Escherichia coli, even though the same combination might be antagonistic against susceptible strains nih.govnih.govresearchgate.netresearchgate.net.
Proposed mechanisms for this synergy in resistant strains include the ability of this compound to enhance the uptake of tetracycline into bacterial cells, potentially by affecting membrane permeability or interfering with efflux pumps that contribute to resistance nih.govresearchgate.netnih.gov. This enhanced uptake can lead to intracellular concentrations of tetracycline sufficient to exert an antibacterial effect, even in strains that are otherwise resistant to tetracycline alone nih.govresearchgate.net. Research has indicated that this compound can enhance the uptake of tetracycline in MDR E. coli strains but not in susceptible strains nih.gov.
The following table summarizes some of the observed in vitro interactions:
| Combination | Bacterial Strain Type | Observed Interaction | Reference(s) |
| This compound + Tetracycline | Susceptible | Antagonism | rxlist.comcabidigitallibrary.orgmicrobiologyresearch.orgnih.gov |
| This compound + Chloramphenicol | Susceptible | Antagonism | rxlist.comoup.comcabidigitallibrary.orgmicrobiologyresearch.orgresearchgate.net |
| This compound + Nitrofurantoin | Susceptible | Antagonism | rxlist.comoup.comrwandafda.gov.rwpharmacy180.comnih.gov |
| This compound + Tetracycline | Multidrug-Resistant | Synergy | nih.govnih.govresearchgate.netresearchgate.net |
Advanced Research Methodologies and Applications
Proteomic Analysis in Resistance Studies
Proteomic analysis plays a significant role in deciphering the complex protein-level changes associated with bacterial resistance to Nalidixic acid. Studies comparing this compound-resistant strains to susceptible counterparts have revealed differential expression of numerous proteins. For instance, proteomic investigations in Escherichia coli resistant to this compound have identified the upregulation of outer membrane proteins such as TolC, OmpT, OmpC, and OmpW, alongside the downregulation of FadL. frontiersin.orgacs.orgacs.orgnih.gov
These findings suggest the involvement of efflux pumps (like those incorporating TolC) and alterations in membrane permeability (mediated by porins like OmpC and OmpF, regulated by systems such as EnvZ/OmpR) as mechanisms contributing to resistance. acs.orgacs.orgnih.gov Other proteomic studies on fluoroquinolone resistance more broadly have also highlighted the role of efflux pumps and changes in major metabolic pathways. frontiersin.orgdovepress.com
Research findings from proteomic studies on this compound resistance in E. coli include:
| Protein | Observed Change in Resistant Strains | Potential Role in Resistance | Source |
| TolC | Upregulated | Efflux pump component | frontiersin.orgacs.orgacs.orgnih.gov |
| OmpT | Upregulated | Outer membrane protein | frontiersin.orgacs.orgacs.orgnih.gov |
| OmpC | Upregulated | Outer membrane protein, Porin | frontiersin.orgacs.orgacs.orgnih.gov |
| OmpW | Upregulated | Outer membrane protein | frontiersin.orgacs.orgacs.orgnih.gov |
| FadL | Downregulated | Outer membrane protein, Fatty acid transport | frontiersin.orgacs.orgacs.orgnih.gov |
| NarG | Low levels observed (in multi-drug resistant strains) | Component of respiratory nitrate reductase | frontiersin.orgdovepress.com |
| NarH | Low levels observed (in multi-drug resistant strains) | Component of respiratory nitrate reductase | frontiersin.orgdovepress.com |
These proteomic insights contribute to a better understanding of the multifactorial nature of antibiotic resistance.
Whole-Genome Sequencing in Resistance Mechanism Elucidation
Whole-Genome Sequencing (WGS) is a powerful tool for identifying the genetic basis of this compound resistance. By sequencing the entire genome of resistant bacterial strains, researchers can pinpoint chromosomal mutations and the presence of acquired genes that confer resistance. nih.govfrontiersin.orgnih.govresearchgate.netasm.orgnih.govasm.orgoup.comoup.comelsevier.esmdpi.com
Key resistance mechanisms elucidated through WGS include mutations in the genes encoding the primary drug targets, DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE). nih.govresearchgate.netasm.orgoup.comoup.comelsevier.es Specific point mutations, such as those at codon Asp87 in gyrA, are frequently associated with high-level this compound resistance. researchgate.netoup.com WGS also facilitates the detection of plasmid-mediated quinolone resistance (PMQR) genes, including qnr variants and aac(6')-1b-cr, which can confer low-level resistance and contribute to the development of higher resistance levels. nih.govasm.orgelsevier.es
WGS allows for the correlation of specific genotypes with observed resistance phenotypes, providing a comprehensive view of resistance mechanisms. nih.govoup.comoup.com Studies have utilized WGS to confirm resistance determinants identified by other methods and to explore the genomic diversity of resistant strains. nih.govfrontiersin.orgnih.govasm.org
Research findings from WGS studies related to this compound resistance include:
| Gene/Element | Type of Alteration | Associated with Resistance to | Notes | Source |
| gyrA | Chromosomal Mutation | This compound, Fluoroquinolones | Often involves specific point mutations (e.g., Asp87) | nih.govfrontiersin.orgresearchgate.netasm.orgoup.comoup.comelsevier.es |
| parC | Chromosomal Mutation | Fluoroquinolones (often with gyrA mutations) | Part of Topoisomerase IV | nih.govasm.orgoup.comoup.comelsevier.es |
| parE | Chromosomal Mutation | Fluoroquinolones (less common than gyrA, parC) | Part of Topoisomerase IV | asm.orgoup.com |
| qnr genes | Plasmid-mediated | Low-level Fluoroquinolone | Protect DNA gyrase | nih.govasm.orgelsevier.esjidc.org |
| aac(6')-1b-cr | Plasmid-mediated | Fluoroquinolones, Aminoglycosides | Modifies ciprofloxacin and other drugs | nih.govasm.orgelsevier.es |
| Single Nucleotide Variations (SNVs) | Chromosomal | This compound | Novel SNVs can be predicted by GWAS/comparative genomics | nih.gov |
Biosensor Reporter Strain Applications
This compound's ability to induce DNA damage by inhibiting DNA gyrase makes it a useful tool in the development and application of bacterial biosensor reporter strains. wikipedia.orgdrugbank.comnih.gov These biosensors are genetically engineered bacteria that produce a measurable signal, such as luminescence or fluorescence, in response to specific environmental stimuli, including genotoxic agents. nih.gov
Bacterial reporter strains designed to detect DNA damage often utilize transcriptional fusions where a promoter responsive to DNA damage (like the recA or sulA promoter, part of the SOS response) is linked to a reporter gene (such as luxCDABE for bioluminescence or gfp for green fluorescent protein). nih.govnih.govcapes.gov.brresearchgate.net this compound is frequently used as a positive control or an inducing agent to validate the functionality and sensitivity of these biosensors. nih.govresearchgate.net Exposure to this compound triggers the DNA damage response, leading to the activation of the promoter and subsequent production of the reporter signal, which can be quantified. nih.govcapes.gov.brresearchgate.net
These biosensors have potential applications in detecting genotoxic compounds in various samples. nih.gov
In Vitro Susceptibility Testing (e.g., Disk Diffusion Method)
In vitro susceptibility testing methods are fundamental for assessing the effectiveness of antibiotics against bacterial isolates. The disk diffusion method, particularly the Kirby-Bauer method, is a widely used and standardized technique. tmmedia.ingtfcc.orgthermofisher.com This method involves placing antibiotic-impregnated disks on an agar plate inoculated with a bacterial culture. tmmedia.ingtfcc.org As the antibiotic diffuses into the agar, it creates a concentration gradient. If the bacteria are susceptible, a clear zone of inhibition forms around the disk where bacterial growth is inhibited. tmmedia.ingtfcc.org The diameter of this zone correlates with the susceptibility of the organism to the antibiotic. tmmedia.in
For this compound, disk diffusion testing is not only used to determine direct susceptibility but also serves as a valuable screening tool for detecting decreased susceptibility or resistance to fluoroquinolone antibiotics, especially those mediated by mutations in DNA gyrase (gyrA). nih.govasm.orgelsevier.esjidc.orggtfcc.orgnih.govasm.org Resistance to this compound, as determined by disk diffusion, can indicate the presence of gyrA mutations that also affect the activity of other fluoroquinolones like ciprofloxacin. nih.govasm.orgelsevier.esjidc.orgnih.govasm.org Standardized procedures and interpretive criteria for this compound disk diffusion are provided by organizations like the Clinical Laboratory Standards Institute (CLSI). tmmedia.ingtfcc.orgnih.govasm.org
This compound disks are commercially available for this purpose. thermofisher.com Studies have evaluated the performance of this compound disks in screening for fluoroquinolone resistance mechanisms in various bacterial species, including Escherichia coli and Neisseria meningitidis. nih.govasm.orgnih.govasm.org
| Method | Application in this compound Research | Key Principle | Source |
| Disk Diffusion | Determining bacterial susceptibility, Screening for fluoroquinolone resistance mechanisms | Measurement of zone of inhibition around antibiotic disk on inoculated agar | nih.govasm.orgelsevier.esjidc.orgtmmedia.ingtfcc.orgthermofisher.comnih.govasm.org |
Q & A
Basic Research Questions
Q. How should researchers design experiments to assess the bactericidal effects of nalidixic acid on Gram-positive and Gram-negative bacteria?
- Methodological Answer : Use standardized bacterial strains (e.g., Bacillus subtilis for Gram-positive, Escherichia coli for Gram-negative) and measure minimum inhibitory concentrations (MICs) via broth microdilution. Include controls for solvent effects (e.g., DMSO). Monitor DNA synthesis inhibition using radiolabeled thymidine incorporation assays and correlate with bactericidal activity via time-kill curves. Morphological changes (e.g., cell elongation, Gram-negative staining in Gram-positive species) should be documented using microscopy .
Q. What experimental protocols ensure the chemical stability of this compound during long-term storage and in aqueous solutions?
- Methodological Answer : Store this compound in sealed, light-protected containers at 4°C to prevent degradation. For aqueous studies, prepare fresh solutions in pH-buffered solvents (e.g., phosphate buffer, pH 7.4) and avoid exposure to strong oxidizers. Validate stability via UV spectrophotometry (peak absorbance at ~260 nm) or HPLC over 24-hour periods .
Q. How can researchers quantify this compound in biological matrices like plasma while minimizing interference from metabolites?
- Methodological Answer : Employ high-performance liquid chromatography (HPLC) with UV detection (260 nm) or gas chromatography (GC) after derivatization. Validate methods using spiked plasma samples and compare retention times with known standards. For metabolite exclusion (e.g., hydroxymethylthis compound), confirm chromatographic separation via tandem mass spectrometry (MS/MS) .
Advanced Research Questions
Q. What mechanistic approaches elucidate the selective inhibition of bacterial DNA synthesis by this compound?
- Methodological Answer : Use subcellular systems (e.g., toluene-treated E. coli lacking DNA polymerase I) to isolate ATP-dependent DNA synthesis. Apply this compound at concentrations near the MIC (e.g., 25 µg/mL) and quantify DNA degradation via spectrophotometric or fluorometric assays. Compare sensitivity of membrane-bound DNA synthesizing systems (e.g., from B. subtilis) to identify target specificity .
Q. How can researchers resolve contradictions in this compound’s reported effects on eukaryotic systems (e.g., lifespan modulation)?
- Methodological Answer : Conduct microdissection assays in model eukaryotes (e.g., Caenorhabditis elegans) using controlled doses (e.g., 10–100 µM) and standardized viability metrics. Pair with genomic analysis (e.g., RNA sequencing) to distinguish direct DNA-targeting effects from off-pathway interactions. Validate findings against known lifespan-altering compounds (e.g., nicotinamide) .
Q. What strategies optimize the sensitivity and reproducibility of this compound quantification in complex biological environments?
- Methodological Answer : Combine HPLC with fluorescence detection (excitation 325 nm, emission 370 nm) for enhanced sensitivity. For reproducibility, adhere to metrological guidelines (e.g., ISO/IEC 17025) for calibration standards and inter-laboratory validation. Cross-validate results using alternative techniques like capillary electrophoresis or immunoassays .
Experimental Design and Data Analysis
Q. How should researchers address variability in this compound’s antibacterial activity across bacterial strains?
- Methodological Answer : Perform dose-response assays across phylogenetically diverse strains (e.g., Pseudomonas aeruginosa, Staphylococcus aureus). Use statistical models (e.g., ANOVA with post-hoc Tukey tests) to analyze MIC variations. Include genetic profiling (e.g., gyrA mutations) to link resistance mechanisms to activity shifts .
Q. What protocols validate the absence of cytotoxic effects when testing this compound in eukaryotic cell cultures?
- Methodological Answer : Conduct parallel assays with mammalian cell lines (e.g., HEK293) using MTT or resazurin-based viability tests. Compare cytotoxicity thresholds (IC50) with antibacterial MICs to establish selectivity indices. Include positive controls (e.g., doxorubicin) and negative controls (culture medium only) .
Retrosynthesis Analysis
AI-Powered Synthesis Planning: Our tool employs the Template_relevance Pistachio, Template_relevance Bkms_metabolic, Template_relevance Pistachio_ringbreaker, Template_relevance Reaxys, Template_relevance Reaxys_biocatalysis model, leveraging a vast database of chemical reactions to predict feasible synthetic routes.
One-Step Synthesis Focus: Specifically designed for one-step synthesis, it provides concise and direct routes for your target compounds, streamlining the synthesis process.
Accurate Predictions: Utilizing the extensive PISTACHIO, BKMS_METABOLIC, PISTACHIO_RINGBREAKER, REAXYS, REAXYS_BIOCATALYSIS database, our tool offers high-accuracy predictions, reflecting the latest in chemical research and data.
Strategy Settings
| Precursor scoring | Relevance Heuristic |
|---|---|
| Min. plausibility | 0.01 |
| Model | Template_relevance |
| Template Set | Pistachio/Bkms_metabolic/Pistachio_ringbreaker/Reaxys/Reaxys_biocatalysis |
| Top-N result to add to graph | 6 |
Feasible Synthetic Routes
Featured Recommendations
| Most viewed | ||
|---|---|---|
| Most popular with customers |
Disclaimer and Information on In-Vitro Research Products
Please be aware that all articles and product information presented on BenchChem are intended solely for informational purposes. The products available for purchase on BenchChem are specifically designed for in-vitro studies, which are conducted outside of living organisms. In-vitro studies, derived from the Latin term "in glass," involve experiments performed in controlled laboratory settings using cells or tissues. It is important to note that these products are not categorized as medicines or drugs, and they have not received approval from the FDA for the prevention, treatment, or cure of any medical condition, ailment, or disease. We must emphasize that any form of bodily introduction of these products into humans or animals is strictly prohibited by law. It is essential to adhere to these guidelines to ensure compliance with legal and ethical standards in research and experimentation.


