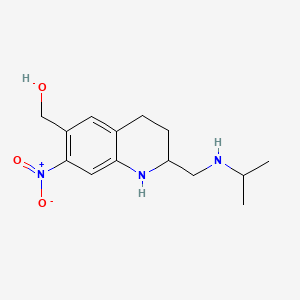
Oxamniquine
Description
Historical Context of Antischistosomal Drug Development
The history of antischistosomal drug development spans several decades, marked by the discovery and implementation of various chemical compounds. Early attempts at treating schistosomiasis involved the use of antimonials, such as potassium antimony tartrate, which were among the first compounds to demonstrate efficacy against the parasite. wikipedia.orgnih.govmims.comcalpaclab.com While these compounds showed activity, they were often associated with significant toxicity and required lengthy treatment regimens. wikipedia.orgnih.gov
The mid-20th century saw the development of other antischistosomal agents, including niridazole and hycanthone. wikipedia.org These drugs represented advances in treatment, but limitations such as side effects and variable efficacy against different Schistosoma species remained challenges. wikipedia.org The advent of Oxamniquine in the 1970s, alongside praziquantel, marked a significant breakthrough, offering more effective and better-tolerated oral treatments. wikipedia.orgwikipedia.org
Evolution of this compound's Role in Schistosomiasis Control Programs
This compound was introduced in the early 1970s and quickly became an important tool in schistosomiasis control programs, particularly in areas endemic for Schistosoma mansoni. wikipedia.orgwikipedia.org It was used extensively, notably in Brazil, for both individual treatment and mass chemotherapy campaigns. wikipedia.orgwikipedia.orgwikipedia.orgwikipedia.org Its effectiveness against the adult worms of S. mansoni contributed to reducing the prevalence and intensity of infection in treated populations. wikipedia.orgontosight.ai
Over time, praziquantel, a broad-spectrum anthelmintic effective against all major Schistosoma species infecting humans (S. mansoni, S. haematobium, and S. japonicum), became the mainstay of mass drug administration programs globally due to its broader efficacy and favorable profile. wikipedia.orgontosight.aiwikipedia.orgamericanelements.comfishersci.cacenmed.com This led to a decrease in the widespread use of this compound in many regions, although it has remained one of the two commercially available drugs effective against S. mansoni. wikipedia.org
The historical use of various drugs highlights the evolving strategies in schistosomiasis control, moving from highly toxic compounds to more targeted and effective chemotherapies.
Current Research Landscape and Unmet Needs in Schistosomiasis Chemotherapy
Despite the success of praziquantel in controlling schistosomiasis-related morbidity, several challenges and unmet needs persist in the area of chemotherapy. A major concern is the heavy reliance on a single drug, praziquantel, for mass treatment, which raises fears of the potential development of widespread drug resistance. wikipedia.orgwikipedia.orgamericanelements.comfishersci.cacenmed.com
This compound's limitation to being effective primarily against Schistosoma mansoni (and S. intercalatum) but not against S. haematobium or S. japonicum presents another challenge in achieving comprehensive control across different endemic regions. wikipedia.orgontosight.aiwikipedia.org Current research is actively exploring the potential of this compound and its derivatives to address these limitations.
Detailed research findings have shed light on the species-specific activity of this compound. Studies have shown that this compound acts as a prodrug that requires enzymatic activation by a schistosome sulfotransferase (SULT) to become an active alkylating agent. wikipedia.orgwikipedia.orgwikipedia.orgfishersci.cauni.lu This activation leads to the covalent binding of the drug to parasite macromolecules, ultimately causing worm paralysis and death. wikipedia.org Research has revealed that the differences in the binding pocket of the SULT enzyme across different Schistosoma species (specifically S. mansoni versus S. haematobium and S. japonicum) explain why this compound is not as effective against S. haematobium and S. japonicum. wikipedia.orgwikipedia.orguni.lu
This understanding of the mechanism of action and the structural differences in the activating enzyme has paved the way for structure-guided drug design efforts. wikipedia.orgwikipedia.orgfishersci.cauni.lu Researchers are developing and testing new this compound derivatives aimed at overcoming the species specificity and achieving broad-spectrum activity against all major human Schistosoma species. wikipedia.orgwikipedia.orgamericanelements.comfishersci.cauni.lu Experimental compounds, such as certain CIDD derivatives, have shown promise in laboratory settings by demonstrating efficacy against S. mansoni, S. haematobium, and S. japonicum. wikipedia.orgamericanelements.comfishersci.ca The goal of this research is to develop a second effective drug with a different mode of action than praziquantel, which could be used in combination therapies to mitigate the risk of resistance and improve treatment outcomes. wikipedia.orgwikipedia.orgamericanelements.comfishersci.ca
The current research landscape is also focused on identifying new drug candidates with novel mechanisms of action and addressing the unmet need for suitable treatments for vulnerable populations, such as preschool-aged children. iiab.mefishersci.ca The investigation into this compound derivatives represents a significant effort to expand the limited arsenal of available antischistosomal drugs and improve control strategies for this persistent neglected tropical disease.
Properties
IUPAC Name |
[7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl]methanol | |
|---|---|---|
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI |
InChI=1S/C14H21N3O3/c1-9(2)15-7-12-4-3-10-5-11(8-18)14(17(19)20)6-13(10)16-12/h5-6,9,12,15-16,18H,3-4,7-8H2,1-2H3 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI Key |
XCGYUJZMCCFSRP-UHFFFAOYSA-N | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Canonical SMILES |
CC(C)NCC1CCC2=CC(=C(C=C2N1)[N+](=O)[O-])CO | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Formula |
C14H21N3O3 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
DSSTOX Substance ID |
DTXSID3023398 | |
| Record name | Oxamniquine | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID3023398 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
Molecular Weight |
279.33 g/mol | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Physical Description |
Solid | |
| Record name | Oxamniquine | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0015228 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Solubility |
1 part in about 3300 parts of water at 27 °C, 1.24e-01 g/L | |
| Record name | Oxamniquine | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB01096 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | OXAMNIQUINE | |
| Source | Hazardous Substances Data Bank (HSDB) | |
| URL | https://pubchem.ncbi.nlm.nih.gov/source/hsdb/6510 | |
| Description | The Hazardous Substances Data Bank (HSDB) is a toxicology database that focuses on the toxicology of potentially hazardous chemicals. It provides information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. The information in HSDB has been assessed by a Scientific Review Panel. | |
| Record name | Oxamniquine | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0015228 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Color/Form |
Pale yellow crystals from isopropanol, Yellow-orange, crystalline solid | |
CAS No. |
21738-42-1, 40247-39-0, 119678-90-9 | |
| Record name | Oxamniquine | |
| Source | CAS Common Chemistry | |
| URL | https://commonchemistry.cas.org/detail?cas_rn=21738-42-1 | |
| Description | CAS Common Chemistry is an open community resource for accessing chemical information. Nearly 500,000 chemical substances from CAS REGISTRY cover areas of community interest, including common and frequently regulated chemicals, and those relevant to high school and undergraduate chemistry classes. This chemical information, curated by our expert scientists, is provided in alignment with our mission as a division of the American Chemical Society. | |
| Explanation | The data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated. | |
| Record name | Oxamniquine [USAN:USP:INN:BAN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0021738421 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Oxamniquine, (+)- | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0040247390 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Oxamniquine, (-)- | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0119678909 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Oxamniquine | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB01096 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | oxamniquine | |
| Source | DTP/NCI | |
| URL | https://dtp.cancer.gov/dtpstandard/servlet/dwindex?searchtype=NSC&outputformat=html&searchlist=352888 | |
| Description | The NCI Development Therapeutics Program (DTP) provides services and resources to the academic and private-sector research communities worldwide to facilitate the discovery and development of new cancer therapeutic agents. | |
| Explanation | Unless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source. | |
| Record name | Oxamniquine | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID3023398 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
| Record name | Oxamniquine | |
| Source | European Chemicals Agency (ECHA) | |
| URL | https://echa.europa.eu/substance-information/-/substanceinfo/100.040.491 | |
| Description | The European Chemicals Agency (ECHA) is an agency of the European Union which is the driving force among regulatory authorities in implementing the EU's groundbreaking chemicals legislation for the benefit of human health and the environment as well as for innovation and competitiveness. | |
| Explanation | Use of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page. | |
| Record name | OXAMNIQUINE | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/0O977R722D | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
| Record name | OXAMNIQUINE, (-)- | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/00BCY677OT | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
| Record name | OXAMNIQUINE, (+)- | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/7GIJ138H3K | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
| Record name | OXAMNIQUINE | |
| Source | Hazardous Substances Data Bank (HSDB) | |
| URL | https://pubchem.ncbi.nlm.nih.gov/source/hsdb/6510 | |
| Description | The Hazardous Substances Data Bank (HSDB) is a toxicology database that focuses on the toxicology of potentially hazardous chemicals. It provides information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. The information in HSDB has been assessed by a Scientific Review Panel. | |
| Record name | Oxamniquine | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0015228 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Melting Point |
147-149 °C, 147 - 149 °C | |
| Record name | Oxamniquine | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB01096 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | OXAMNIQUINE | |
| Source | Hazardous Substances Data Bank (HSDB) | |
| URL | https://pubchem.ncbi.nlm.nih.gov/source/hsdb/6510 | |
| Description | The Hazardous Substances Data Bank (HSDB) is a toxicology database that focuses on the toxicology of potentially hazardous chemicals. It provides information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. The information in HSDB has been assessed by a Scientific Review Panel. | |
| Record name | Oxamniquine | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0015228 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Mechanisms and Epidemiology of Oxamniquine Resistance
Genetic Basis of Resistance Development
The development of resistance to oxamniquine in S. mansoni is primarily driven by genetic changes within the parasite.
Identification of Resistance as a Single Autosomal Recessive Trait
Research has established that this compound resistance in Schistosoma mansoni is controlled by a single autosomal recessive gene. nih.govcambridge.orgresearchgate.netnih.gov This was demonstrated through genetic crosses between drug-sensitive and drug-resistant schistosomes. cambridge.orgresearchgate.netnih.gov Studies involving F1 and F2 generations, as well as backcrosses, consistently showed that the resistance phenotype is only expressed when a parasite is homozygous for the resistance alleles. nih.govresearchgate.net Resistance confers a significant reduction in drug sensitivity, in some cases up to ~500-fold. nih.gov
Role of Loss-of-Function Mutations in Sulfotransferase Encoding Genes
This compound is administered as a prodrug and requires metabolic activation by a schistosome sulfotransferase (SmSULT-OR) to become effective. nih.govcambridge.orgplos.orgscispace.complos.org The gene encoding this enzyme, SmSULT-OR, is located on chromosome 6 of S. mansoni. scispace.com Resistance to this compound is primarily caused by loss-of-function mutations within the SmSULT-OR gene. nih.govplos.orgscispace.complos.orgnih.govnih.gov These mutations result in a defective sulfotransferase enzyme, which is unable to efficiently activate the this compound prodrug. nih.govplos.orgscispace.comnih.gov Several types of mutations have been identified, including non-synonymous single nucleotide polymorphisms (SNPs), deletions, duplications, and premature stop codons, all of which can disrupt the function of SmSULT-OR. nih.govnih.govnih.gov For instance, the p.E142del and p.C35R mutations have been identified as causing resistance by disrupting the active site of the enzyme. nih.govplos.org
Epidemiological Studies of Resistance Emergence and Distribution in Endemic Regions
Epidemiological studies have provided insights into the emergence and distribution of this compound resistance in areas where schistosomiasis is endemic. Resistant parasites were detected in Brazil in the 1970s, even before widespread mass drug administration programs. nih.govplos.org Similar observations of poor treatment response in patients were made in East Africa. nih.govplos.org Experimental infections in mice using parasites isolated from patients with poor treatment outcomes confirmed the existence of this compound-resistant parasites in regions like Brazil and Kenya. nih.govresearchgate.netplos.orgscielo.br
Importantly, studies have revealed that this compound resistance alleles were already widespread in Old World S. mansoni populations, predating the extensive deployment of the drug in the 1970s. nih.govplos.orgplos.orgnih.govifremer.frbiorxiv.orgajtmh.org Analysis of parasite populations from areas with minimal this compound usage, such as parts of Africa and the Middle East, showed notable frequencies of resistance alleles. nih.govplos.orgbiorxiv.org For example, studies found this compound resistance allele frequencies ranging from 4.29% in the Middle East to 14.91% in East African parasite populations. nih.govplos.orgbiorxiv.org The presence of the p.E142del allele in both West Africa and Puerto Rico with identical flanking SNPs suggests that resistance alleles were transported to the New World with S. mansoni during the transatlantic slave trade, further indicating that these alleles existed long before this compound was used therapeutically. nih.govplos.orgifremer.frbiorxiv.org This widespread standing variation for resistance has significant implications for how rapidly resistance can spread under drug pressure. nih.govplos.orgplos.org
Data on the frequency of this compound resistance alleles in different Old World populations:
| Region | This compound Resistance Allele Frequency |
| Middle East | 4.29% |
| East Africa | 14.91% |
| West Africa | 4.29% |
Two specific resistance mutations, p.W120R and p.N171IfsX28, were found to be particularly common (>5%) in East African and Middle-Eastern populations. nih.govbiorxiv.org
Molecular Surveillance and Characterization of Resistance Alleles
Molecular surveillance plays a vital role in monitoring the prevalence and distribution of this compound resistance alleles in parasite populations. nih.govifremer.fr By sequencing the SmSULT-OR gene from parasite samples collected in endemic areas, researchers can identify and characterize existing and emerging resistance mutations. nih.govnih.govnih.gov Studies have involved sequencing SmSULT-OR from individual miracidia obtained from infected patients. nih.gov This approach has led to the identification of various mutations, including previously known ones like p.E142del and p.C35R, as well as novel variants predicted to cause loss-of-function in the sulfotransferase. nih.govnih.govnih.gov
Functional validation of identified mutations is crucial, often involving the expression of recombinant sulfotransferase proteins with the identified mutations and testing their ability to activate this compound in vitro. nih.govnih.govplos.org This helps confirm which genetic variants truly confer resistance. Molecular surveillance efforts, such as those involving large parasite collections, are valuable for regularly updating the understanding of the resistance landscape in different endemic regions. nih.govplos.org However, it is important to pair molecular screening with functional evaluation or computational prediction to determine the phenotypic impact of identified mutations. nih.govplos.org
Research Strategies to Overcome this compound Resistance
Addressing this compound resistance involves several research strategies. One approach focuses on developing new this compound derivatives that can overcome the limitations of the parent compound, including resistance. plos.orgplos.orgnih.govresearchgate.net Guided by structural data of the SmSULT-OR enzyme and its interaction with this compound, researchers are designing and synthesizing novel derivatives. plos.orgnih.govresearchgate.net These efforts aim to create compounds that can be activated by both wild-type and mutated sulfotransferase enzymes or that have alternative mechanisms of action. plos.orgnih.gov Studies have identified promising this compound derivatives with potent activity against S. mansoni, including resistant strains, and even against other Schistosoma species that are not susceptible to the original this compound. plos.orgplos.orgnih.govresearchgate.net For example, derivatives like CIDD-0150610 and CIDD-0150303 have shown high killing rates against S. mansoni, S. haematobium, and S. japonicum in vitro. nih.govresearchgate.net In vivo studies with these derivatives have also demonstrated significant worm burden reductions. nih.govresearchgate.net
Another strategy involves exploring combination therapies, where this compound or its derivatives could be used alongside other antischistosomal drugs like praziquantel, particularly against resistant strains. nih.gov Research into the molecular mechanisms of resistance and the structure of the activating enzyme also provides a framework for rational drug design. scispace.complos.org Furthermore, understanding the population genetics of resistance alleles and their distribution through molecular surveillance can inform treatment strategies and help manage the spread of resistance. nih.govifremer.frbiorxiv.orgfrontiersin.org Advanced genetic tools, such as CRISPR-Cas systems, are also being explored to understand the function of genes related to drug resistance, which could potentially aid in developing new interventions. wellcomeopenresearch.org
Development of Next-Generation Derivatives with Modified Activation or Target Profiles
The understanding of this compound's mechanism of action and the basis of resistance, particularly the role of the SmSULT-OR enzyme, has provided a framework for the rational design of new this compound derivatives. plos.orgscispace.comunizik.edu.ng The goal of developing next-generation derivatives is to create compounds with improved efficacy, broader activity against different Schistosoma species (including S. haematobium and S. japonicum, which are not effectively treated by the original this compound), and reduced potential for resistance. plos.orgresearchgate.netresearchgate.netbiorxiv.org
Structure-based drug design approaches, guided by X-ray crystallographic data of sulfotransferase enzymes and structure-activity relationship (SAR) studies, have been employed to design and synthesize hundreds of this compound derivatives. plos.orgresearchgate.netbiorxiv.org These efforts aim to modify the chemical structure of this compound to enhance its interaction with the target sulfotransferase or to be activated by alternative pathways or target different molecules within the parasite.
Research has identified promising new derivatives with potent in vitro killing activity against multiple Schistosoma species. plos.orgresearchgate.netbiorxiv.org For example, compounds like CIDD-0066790, CIDD-0072229 (the R-enantiomer of CIDD-0066790), CIDD-0149830, CIDD-0150610, and CIDD-0150303 have shown broad-species activity, killing S. mansoni, S. haematobium, and S. japonicum in laboratory settings. plos.orgresearchgate.netbiorxiv.org
Studies suggest that the differential activity of these derivatives against various Schistosoma species is related to their ability to fit and be efficiently sulfated within the binding pockets of the respective sulfotransferases (SmSULT-OR, ShSULT-OR, and SjSULT-OR). plos.orgresearchgate.netresearchgate.net Molecular simulations and crystallographic studies of these derivatives bound to sulfotransferases provide insights into the structural mechanisms underlying their efficacy and selectivity. researchgate.netbiorxiv.orgresearchgate.net
Some derivatives have demonstrated rapid killing times in vitro. For instance, CIDD-0149830 has been shown to kill 100% of S. mansoni, S. haematobium, and S. japonicum adult worms within 7 days in vitro. plos.orgresearchgate.net Other derivatives, like CIDD-0150610 and CIDD-0150303, have also shown 100% killing of S. mansoni within 24 hours at certain concentrations in vitro. biorxiv.orgresearchgate.net
Furthermore, some of these next-generation derivatives have shown efficacy against immature schistosome stages, a limitation of praziquantel. biorxiv.org For example, CIDD-0150303 demonstrated 100% killing for all life stages of S. mansoni in vitro and effective reduction in worm burden against immature stages in vivo in animal models. biorxiv.org
Here is a table summarizing the in vitro killing activity of some promising this compound derivatives against S. mansoni:
| Compound | Concentration (µM) | % Killing (S. mansoni) | Time (Days) | Source |
| This compound | 71.5 | 40 | 14 | researchgate.net |
| CIDD-0149830 | 143 | 100 | 7 | plos.orgresearchgate.net |
| CIDD-0072229 | Not specified | Better than parent | Not specified | plos.org |
| CIDD-0150610 | 143 | 100 | <1 | biorxiv.orgresearchgate.net |
| CIDD-0150303 | 143 | 100 | <1 | biorxiv.orgresearchgate.net |
Note: Killing times and concentrations may vary depending on specific experimental conditions.
Investigation of Combination Therapy Approaches to Mitigate Resistance Evolution
The emergence of drug resistance, particularly to praziquantel which is currently the sole drug recommended for mass drug administration (MDA), highlights the urgent need for alternative treatment strategies, including combination therapies. plos.orgresearchgate.netplos.org Using drugs with different mechanisms of action in combination can help mitigate the evolution of resistance by requiring parasites to develop resistance to multiple drugs simultaneously, a less likely evolutionary event. oup.com
Given the history of this compound use and the identified mechanism of resistance distinct from that of praziquantel, investigating combination therapies involving this compound or its derivatives is a logical step. ajtmh.org While this compound resistance is linked to the sulfotransferase enzyme, the molecular basis of praziquantel resistance is still being investigated but appears to involve different mechanisms, possibly related to calcium channels like SmTRPMPZQ. frontiersin.org
The development of next-generation this compound derivatives with broader species coverage and activity against different life stages makes them attractive candidates for combination therapy, potentially alongside praziquantel. plos.orgresearchgate.netbiorxiv.orgresearchgate.net The aim is to leverage the distinct targets and activation pathways of the combined drugs to achieve higher cure rates, prevent the selection of resistant parasites, and potentially reduce treatment durations or dosages.
Pre-clinical studies in animal models have investigated the efficacy of combining praziquantel with the newly developed this compound derivatives. For instance, treatment with a single dose of praziquantel in combination with CIDD-0150303 reduced the worm burden of a praziquantel-resistant parasite strain in mice. biorxiv.orgresearchgate.net This suggests that combining drugs with different mechanisms of action can be effective against resistant parasites and could help overcome the limitations of monotherapy.
The investigation of combination therapy approaches is crucial for the long-term sustainability of schistosomiasis control programs, especially in the face of rising concerns about drug resistance. plos.orgplos.org Developing and implementing effective combination therapies could provide a more robust strategy to combat existing resistance and slow down the evolution of new resistant strains.
Pharmacological Research and Drug Disposition Dynamics of Oxamniquine
Elucidation of Bioactivation Pathways and Host Enzyme Interactions
Oxamniquine functions as a prodrug, requiring enzymatic activation within the Schistosoma worm to exert its schistosomicidal effects. Evidence strongly suggests that a schistosome-specific sulfotransferase enzyme is responsible for this activation. This enzyme is notably absent in drug-resistant parasites. The activation process is hypothesized to involve the conversion of this compound to a reactive ester, likely a sulfate. nih.govresearchgate.netscielo.brnih.govnih.gov
Studies have provided data supporting the role of a sulfotransferase. For instance, parasite extracts lose their activating capability upon dialysis, indicating the requirement of a dialyzable cofactor. nih.govresearchgate.netscielo.br The addition of 3'-phosphoadenosine 5'-phosphosulfate (PAPS), a known sulfate donor, restored the activity of the dialyzate, strongly suggesting the involvement of a sulfotransferase. nih.govresearchgate.netscielo.br Furthermore, classical sulfotransferase substrates such as beta-estradiol and quercetin competitively inhibited the activation of this compound in vitro. nih.govresearchgate.netscielo.br These substrates could also be sulfonated in vitro using extracts from sensitive, but not resistant, schistosomes. nih.govresearchgate.netscielo.br
Gel filtration analysis of the activating factor from sensitive schistosomes revealed an elution profile corresponding to a molecular mass of approximately 32 kDa, consistent with the typical size of sulfotransferase subunits. nih.govresearchgate.netscielo.br Ion exchange and affinity chromatography further confirmed the sulfotransferase nature of the enzyme. nih.govresearchgate.netscielo.br
This selective activation by a parasite enzyme, specifically S. mansoni sulfotransferase (SmSULT-OR), allows for selective toxicity to the worm while minimizing effects on human enzymes. nih.govresearchgate.netfrancis-press.com Structural data, including X-ray crystallography, of the S. mansoni sulfotransferase have guided the development of this compound derivatives aimed at expanding efficacy to other Schistosoma species like S. haematobium and S. japonicum. plos.orgnih.gov
Pharmacokinetic/Pharmacodynamic (PK/PD) Relationship Modeling relevant to Schistosome Biology
Examination of the PK/PD relationship for this compound has revealed an in vitro-in vivo paradox. Maximal clinical plasma concentrations were found to be significantly lower (five to ten times) than the concentrations required for efficacious in vitro schistosomal killing. nih.govresearchgate.netresearchgate.netnih.govresearchgate.net This paradox is particularly relevant due to the parasite's residence in the vasculature between the intestine and the liver. nih.govresearchgate.netnih.govresearchgate.net
Resolution of In Vitro-In Vivo Efficacy Paradoxes through Physiological Concentration Modeling (e.g., Portal Vein Concentrations)
Modeling of pharmacokinetic data to determine portal vein concentrations has helped to resolve the in vitro-in vivo paradox. nih.govresearchgate.netnih.govresearchgate.net These modeled portal concentrations align better with the in vitro efficacy studies and provide a rationale for the required human dose. nih.govresearchgate.netnih.govresearchgate.net The concept is that the parasite, residing in the portal circulation, is exposed to higher concentrations of the drug before it is extensively metabolized or cleared systemically. nih.govresearchgate.netnih.govresearchgate.net Achieving high portal concentrations is facilitated by rapid oral absorption, favorable solubility, permeability, and minimal intestinal metabolism. researchgate.netnih.gov
Predictive Modeling of Drug Exposure in Preclinical Animal Models
In silico models have been employed to predict murine dosing regimens that would recapitulate the portal concentration and time course observed in humans. researchgate.netresearchgate.netnih.govresearchgate.net Follow-up pharmacokinetic studies in mice have verified that oral gavage doses of 50-100 mg/kg of this compound formulated in acetate buffer can indeed replicate the exposure achieved with the 20-40 mg/kg dose typically used in patients. researchgate.netresearchgate.netnih.govresearchgate.net This modeling and validation in preclinical models are crucial for translating in vitro findings to in vivo efficacy studies and for the development of new this compound analogs. researchgate.netresearchgate.netnih.gov
Metabolic Fate and Excretion Mechanisms in Mammalian Hosts
In mammalian hosts, this compound undergoes extensive metabolism, primarily in the liver, to inactive metabolites. wikipedia.orgdrugbank.com The major metabolite is the 6-carboxy derivative, which is subsequently excreted, primarily in the urine. wikipedia.org About 70% of an administered dose of this compound is excreted as the 6-carboxy metabolite within 12 hours. wikipedia.org Traces of the 2-carboxy metabolite have also been detected in urine. wikipedia.org this compound is rapidly cleared, with a plasma half-life typically ranging from 1 to 2.5 hours. wikipedia.orgdrugbank.com Studies in dogs have suggested the occurrence of first-pass metabolism in the gut wall. who.int Excretion also occurs via bile. researchgate.netnih.govresearchgate.net
Research on Potential Drug-Drug Interactions involving this compound and other Xenobiotics (e.g., Enzyme Inhibition/Induction)
Research on potential drug-drug interactions involving this compound has indicated that it may decrease the metabolism of several other compounds. drugbank.com This suggests a potential for enzyme inhibition. For example, the metabolism of amoxapine, amphetamine, amprenavir, antipyrine, arformoterol, aripiprazole, aripiprazole lauroxil, diltiazem, diphenhydramine, dolasetron, domperidone, donepezil, doxazosin, doxepin, istradefylline, labetalol, levobetaxolol, lidocaine, lisuride, and lofexidine may be decreased when combined with this compound. drugbank.com Additionally, the serum concentration of doxorubicin and dextroamphetamine (an active metabolite of lisdexamfetamine) can be increased when used in combination with this compound. drugbank.com These findings highlight the potential for pharmacokinetic interactions where this compound may inhibit the metabolism of co-administered drugs.
Drug Discovery and Preclinical Development of Oxamniquine-inspired Compounds
Synthetic Methodologies and Chemical Derivatization Strategies
The synthesis of oxamniquine derivatives has been a key aspect of developing new antischistosomal agents. An iterative process involving design, synthesis, and testing has been employed to identify promising candidates. nih.govplos.orgplos.orgresearchgate.net Chemical derivatization strategies have focused on modifying the this compound structure to enhance its activity and overcome limitations. nih.govnih.gov For instance, the Mannich reaction has been utilized to synthesize new this compound derivatives, resulting in unexpected cyclized and etherified products. nih.gov Studies on structure-activity relationships (SAR) have indicated that modifications to the side alkylamine group of this compound can maintain activity, although this may be associated with increased toxicity. nih.gov Over 350 this compound derivatives have been designed, synthesized, and tested. plos.orgresearchgate.netdatadryad.orgnih.govbiorxiv.org
Application of Structure-Based Drug Design (SBDD) and X-ray Crystallography in Analog Development
Structure-Based Drug Design (SBDD) has played a crucial role in the development of this compound analogs. nih.govplos.orgplos.orgresearchgate.netosti.govresearchgate.netacs.org This approach utilizes structural data, particularly from X-ray crystallography, to guide the design of new compounds. nih.govplos.orgplos.orgresearchgate.netosti.govresearchgate.netacs.org The identification of the Schistosoma mansoni sulfotransferase (SmSULT-OR) as the enzyme responsible for activating this compound has been pivotal. nih.govresearchgate.netresearchgate.netresearchgate.netplos.org X-ray crystallographic studies of SmSULT-OR, sometimes co-crystallized with this compound or its analogs, have provided insights into the drug's mode of action and how derivatives fit into the enzyme's binding pocket. nih.govplos.orgplos.orgresearchgate.netresearchgate.netdatadryad.orgbiorxiv.orgosti.govresearchgate.net This structural information has facilitated the rational design of derivatives aimed at improving binding interactions and efficacy against different schistosome species, including S. haematobium and S. japonicum, which are less susceptible to the parent compound. nih.govplos.orgresearchgate.netresearchgate.net For example, X-ray crystal structures of potent derivatives like CIDD-0150303 and CIDD-0150610 have shown that the SULT active site can accommodate modifications, guiding further optimization. plos.orgresearchgate.netdatadryad.orgbiorxiv.org
Utilization of Computational Chemistry and Molecular Simulations for Lead Optimization
Computational chemistry and molecular simulations have been employed to support the lead optimization process for this compound derivatives. gassergroup.comresearchgate.netnih.gov In silico models have been used to predict murine dosing that would recapitulate human pharmacokinetic conditions, specifically focusing on portal vein concentrations where S. mansoni resides. nih.govnih.govresearchgate.net Molecular dynamics simulations have provided insights into the structural dynamics induced by this compound binding to sulfotransferases, helping to understand the species-specific efficacy observed with the parent drug. researchgate.netgassergroup.comresearchgate.netnih.gov These computational approaches support the in vitro findings and aid in understanding the interaction between the drug candidates and their target proteins. gassergroup.comresearchgate.netnih.gov
In Vitro Efficacy Assessment in Defined Schistosome Models
In vitro efficacy assessment is a critical step in the preclinical evaluation of this compound-inspired compounds. These studies typically involve exposing schistosome parasites to different concentrations of the drug candidates under controlled laboratory conditions. plos.orgplos.orgresearchgate.netdatadryad.orgplos.org
Comprehensive Killing Assays Across Various Schistosome Life Stages (Adult Worms, Immature Forms)
In vitro killing assays are conducted to evaluate the efficacy of this compound derivatives against various life stages of Schistosoma species, including adult worms (males and females) and immature forms (schistosomula). plos.orgplos.orgresearchgate.netdatadryad.orgresearchgate.netresearchgate.netplos.org These assays help determine the concentration required to achieve a certain level of parasite killing. For instance, studies have shown that certain derivatives can achieve 100% killing of adult S. mansoni, S. haematobium, and S. japonicum worms within a few days at specific concentrations. nih.govplos.orgplos.orgresearchgate.netdatadryad.orgnih.govbiorxiv.orgplos.orgbioworld.com Some compounds, like CIDD-0150303, have demonstrated 100% killing across all life stages tested in vitro at a concentration of 143 μM. plos.orgdatadryad.orgnih.govbiorxiv.orgresearchgate.netbioworld.com
Data from in vitro screens highlight the varying efficacy of different derivatives against the three major human schistosome species. For example, CIDD-0149830 has shown rapid and complete killing against all three species in vitro. nih.govplos.orgresearchgate.netplos.org
Evaluation of Efficacy against Phenotypically and Genetically Resistant Schistosome Strains
Evaluating the efficacy of this compound derivatives against resistant schistosome strains is crucial due to the emergence of drug resistance. plos.orgresearchgate.netplos.orgscielo.brscite.aibioline.org.br Resistance to this compound in S. mansoni is linked to loss-of-function mutations in the SmSULT-OR enzyme. researchgate.netplos.org Studies have investigated the activity of new derivatives against strains known to be resistant to this compound. plos.orgplos.orgscielo.brscite.aibioline.org.br Some novel compounds have demonstrated the ability to kill praziquantel-resistant parasites in animal models when used in combination with praziquantel. plos.orgresearchgate.netdatadryad.orgnih.govbiorxiv.orgbioworld.com This suggests that this compound-inspired compounds with a different mechanism of action could be valuable in combination therapies to combat existing or emerging resistance. plos.orgresearchgate.netosti.govplos.org
In Vivo Efficacy Studies in Laboratory Animal Models (e.g., Murine and Hamster Models of Schistosomiasis)
In vivo studies using laboratory animal models, such as mice and hamsters, are essential to assess the efficacy of this compound derivatives in a living system. plos.orgdatadryad.orgnih.govbiorxiv.orgresearchgate.netplos.orggassergroup.comresearchgate.netnih.govbioworld.comscielo.brscite.aibioline.org.brscielo.brmdpi.com These models allow for the evaluation of worm burden reduction and the assessment of efficacy against different schistosome species and life stages in a more complex environment. plos.orgdatadryad.orgnih.govbiorxiv.orgresearchgate.netgassergroup.comresearchgate.netnih.govbioworld.comscielo.brscite.aibioline.org.brscielo.br
Studies in mice infected with S. mansoni, S. haematobium, and S. japonicum have been conducted to evaluate the efficacy of lead this compound derivatives. plos.orgdatadryad.orgnih.govbiorxiv.orgresearchgate.netbioworld.com Worm burden reduction is a key metric in these in vivo studies. plos.orgdatadryad.orgnih.govbiorxiv.orgresearchgate.netbioworld.comscielo.brscite.ai For example, specific derivatives have shown significant worm burden reductions against S. mansoni, S. haematobium, and S. japonicum in mouse models. plos.orgdatadryad.orgnih.govbiorxiv.orgresearchgate.netbioworld.com
Studies have also evaluated the efficacy of these compounds against immature schistosome stages in vivo, as praziquantel is less effective against these forms. plos.orgdatadryad.orgnih.govbiorxiv.orgresearchgate.netbioworld.com For example, CIDD-0150303 demonstrated effective reduction in juvenile worm burden against S. mansoni in vivo. plos.orgdatadryad.orgnih.govbiorxiv.orgresearchgate.netbioworld.com
Quantitative Assessment of Worm Burden Reduction
Preclinical studies evaluating this compound-inspired compounds often utilize in vivo models, typically in mice or hamsters, infected with Schistosoma species. The primary endpoint for assessing efficacy is the quantitative reduction in worm burden compared to untreated control groups.
Studies have investigated the efficacy of various this compound derivatives. For instance, a study involving hamsters infected with S. mansoni showed that treatment with CIDD-0150303 at 100 mg/kg by oral gavage resulted in an 81.8% reduction in worm burden. CIDD-0149830 showed an 80.2% reduction against S. haematobium, and CIDD-066790 achieved an 86.7% reduction against S. japonicum at the same dose and route plos.orgresearchgate.net. In comparison, this compound itself reduced S. mansoni worm burden by 93% in hamsters plos.org.
Further in vivo studies in mice infected with juvenile S. mansoni demonstrated that CIDD-0150303 at 100 mg/kg significantly reduced worm burden when administered at different time points post-infection. Reductions of 63.8%, 48.9%, and 54.1% were observed when treated on days 25, 28, and 32 post-infection, respectively plos.org.
Combination therapy with this compound-inspired compounds and praziquantel (PZQ) has also been explored to address PZQ resistance. In an animal model using a PZQ-resistant S. mansoni strain, treatment with a combination of PZQ and CIDD-0150303 (100 mg/kg of each) significantly reduced the worm burden by 90.8% plos.orgresearchgate.net. This suggests that certain this compound derivatives can be effective against resistant strains and may be valuable in combination therapies.
Some studies have also investigated polymeric prodrugs of this compound. However, efficacy trials with certain biopolymers derived from this compound did not demonstrate better effects on reducing worm burden than this compound itself nih.gov.
The following table summarizes some preclinical worm burden reduction data for selected this compound derivatives:
| Compound | Schistosoma Species | Host | Dose (mg/kg) | Route | Worm Burden Reduction (%) | Citation |
| CIDD-0150303 | S. mansoni | Hamster | 100 | Oral | 81.8 | plos.orgresearchgate.net |
| CIDD-0149830 | S. haematobium | Hamster | 100 | Oral | 80.2 | plos.orgresearchgate.net |
| CIDD-066790 | S. japonicum | Hamster | 100 | Oral | 86.7 | plos.orgresearchgate.net |
| CIDD-0150303 | S. mansoni (Juvenile) | Mouse | 100 | Oral | 63.8 (25 dpi) | plos.org |
| CIDD-0150303 | S. mansoni (Juvenile) | Mouse | 100 | Oral | 48.9 (28 dpi) | plos.org |
| CIDD-0150303 | S. mansoni (Juvenile) | Mouse | 100 | Oral | 54.1 (32 dpi) | plos.org |
| PZQ + CIDD-0150303 | S. mansoni (PZQ-R) | Mouse | 100 + 100 | Oral | 90.8 | plos.orgresearchgate.net |
| This compound | S. mansoni | Hamster | 100 | Oral | 93 | plos.org |
Evaluation of Impact on Parasite Fecundity and Egg Viability
Beyond reducing worm burden, the impact of antischistosomal compounds on parasite fecundity (egg production) and egg viability is a critical aspect of preclinical evaluation. Reducing the number of viable eggs is essential for interrupting the parasite's life cycle and preventing the transmission of schistosomiasis.
This compound is known to cause male worms to shift to the liver, and while female worms may return to the mesentery, they can no longer release eggs effectively smolecule.comncats.io. This disruption of worm pairing and location indirectly impacts egg production.
Further detailed investigations into the specific effects of novel this compound-inspired compounds on the reproductive capacity and egg viability of susceptible and resistant Schistosoma strains at various developmental stages are crucial during preclinical development.
Research into Novel Formulation and Drug Delivery Systems for Optimized Pharmacokinetic Profiles and Stability
Optimizing the pharmacokinetic profile and stability of this compound and its derivatives is a key area of preclinical research to improve efficacy, reduce dosing frequency, and potentially mitigate limitations such as poor solubility or rapid metabolism. Novel formulation and drug delivery systems are being investigated to achieve these goals.
One approach involves the development of prodrugs, where the parent compound is chemically modified to improve its absorption, distribution, metabolism, and excretion (ADME) properties. Studies have explored this compound prodrugs, including polymeric forms, with the aim of achieving prolonged biological action and improved bioavailability researchgate.netakjournals.com. While some early polymeric prodrugs did not show improved worm burden reduction compared to this compound, the concept of using prodrug strategies to enhance pharmacokinetic profiles remains an active area of research nih.gov.
Liposomal encapsulation is another delivery system being investigated. Liposome-entrapped this compound (LOXA) has been tested in murine models of S. mansoni infection. LOXA administered subcutaneously demonstrated a significant reduction in worm burden compared to free this compound, particularly when given around the time of infection. Maximum efficacy, with a 97% reduction in parasite number, was observed with subcutaneous LOXA administered one day before infection scielo.br. This suggests that liposomal formulations can influence the delivery and efficacy of this compound.
Research also focuses on understanding the pharmacokinetic/pharmacodynamic (PK/PD) relationship of this compound and its analogs to inform formulation development. Studies have identified an in vitro-in vivo paradox with this compound, where maximal clinical plasma concentrations are significantly lower than the in vitro concentrations required for worm killing nih.govresearchgate.net. Modeling suggests that the parasite's location in the portal vasculature means that portal concentrations are more relevant to efficacy than systemic plasma concentrations nih.govresearchgate.net. This understanding guides the design of formulations aimed at achieving higher and sustained concentrations in the portal circulation.
Improving physicochemical properties like solubility and permeability is also crucial for optimizing oral absorption and achieving favorable portal concentrations nih.govresearchgate.net. Novel this compound analogs are being designed with optimized properties to enhance oral bioavailability and improve in vivo activity nih.gov.
Additionally, research into novel oral formulations, such as floating tablets, has been explored to potentially prolong gastric residence time and improve drug absorption .
Compound Names and PubChem CIDs
| Compound Name | PubChem CID |
| This compound | 21738-42-1 |
| Praziquantel | 5000 |
| CIDD-0150610 | - |
| CIDD-0150303 | - |
| CIDD-0149830 | - |
| CIDD-066790 | - |
| CIDD-0072229 | - |
| CIDD-007229 | - |
| CIDD-0066790 | - |
| Fc-CH2-OXA | - |
| Rc-CH2-OXA | - |
| Bn-CH2-OXA | - |
This compound (OXA) is a well-established antischistosomal drug, historically used against Schistosoma mansoni. Its mechanism involves activation by a schistosome sulfotransferase (SULT), leading to parasite death. While effective against S. mansoni, OXA has limitations, including inactivity against S. haematobium and S. japonicum, and the emergence of resistance. This has driven research into developing novel this compound-inspired compounds with broader activity and improved properties.
Quantitative Assessment of Worm Burden Reduction
Preclinical studies evaluating this compound-inspired compounds often utilize in vivo models, typically in mice or hamsters, infected with Schistosoma species. The primary endpoint for assessing efficacy is the quantitative reduction in worm burden compared to untreated control groups.
Studies have investigated the efficacy of various this compound derivatives. For instance, a study involving hamsters infected with S. mansoni showed that treatment with CIDD-0150303 at 100 mg/kg by oral gavage resulted in an 81.8% reduction in worm burden. CIDD-0149830 showed an 80.2% reduction against S. haematobium, and CIDD-066790 achieved an 86.7% reduction against S. japonicum at the same dose and route. plos.orgresearchgate.net In comparison, this compound itself reduced S. mansoni worm burden by 93% in hamsters. plos.org
Further in vivo studies in mice infected with juvenile S. mansoni demonstrated that CIDD-0150303 at 100 mg/kg significantly reduced worm burden when administered at different time points post-infection. Reductions of 63.8%, 48.9%, and 54.1% were observed when treated on days 25, 28, and 32 post-infection, respectively. plos.org
Combination therapy with this compound-inspired compounds and praziquantel (PZQ) has also been explored to address PZQ resistance. In an animal model using a PZQ-resistant S. mansoni strain, treatment with a combination of PZQ and CIDD-0150303 (100 mg/kg of each) significantly reduced the worm burden by 90.8%. plos.orgresearchgate.net This suggests that certain this compound derivatives can be effective against resistant strains and may be valuable in combination therapies.
Some studies have also investigated polymeric prodrugs of this compound. However, efficacy trials with certain biopolymers derived from this compound did not demonstrate better effects on reducing worm burden than this compound itself. nih.gov
The following table summarizes some preclinical worm burden reduction data for selected this compound derivatives:
| Compound | Schistosoma Species | Host | Dose (mg/kg) | Route | Worm Burden Reduction (%) | Citation |
| CIDD-0150303 | S. mansoni | Hamster | 100 | Oral | 81.8 | plos.orgresearchgate.net |
| CIDD-0149830 | S. haematobium | Hamster | 100 | Oral | 80.2 | plos.orgresearchgate.net |
| CIDD-066790 | S. japonicum | Hamster | 100 | Oral | 86.7 | plos.orgresearchgate.net |
| CIDD-0150303 | S. mansoni (Juvenile) | Mouse | 100 | Oral | 63.8 (25 dpi) | plos.org |
| CIDD-0150303 | S. mansoni (Juvenile) | Mouse | 100 | Oral | 48.9 (28 dpi) | plos.org |
| CIDD-0150303 | S. mansoni (Juvenile) | Mouse | 100 | Oral | 54.1 (32 dpi) | plos.org |
| PZQ + CIDD-0150303 | S. mansoni (PZQ-R) | Mouse | 100 + 100 | Oral | 90.8 | plos.orgresearchgate.net |
| This compound | S. mansoni | Hamster | 100 | Oral | 93 | plos.org |
Evaluation of Impact on Parasite Fecundity and Egg Viability
Beyond reducing worm burden, the impact of antischistosomal compounds on parasite fecundity (egg production) and egg viability is a critical aspect of preclinical evaluation. Reducing the number of viable eggs is essential for interrupting the parasite's life cycle and preventing the transmission of schistosomiasis.
This compound is known to cause male worms to shift to the liver, and while female worms may return to the mesentery, they can no longer release eggs effectively. smolecule.comncats.io This disruption of worm pairing and location indirectly impacts egg production.
Further detailed investigations into the specific effects of novel this compound-inspired compounds on the reproductive capacity and egg viability of susceptible and resistant Schistosoma strains at various developmental stages are crucial during preclinical development.
Research into Novel Formulation and Drug Delivery Systems for Optimized Pharmacokinetic Profiles and Stability
Optimizing the pharmacokinetic profile and stability of this compound and its derivatives is a key area of preclinical research to improve efficacy, reduce dosing frequency, and potentially mitigate limitations such as poor solubility or rapid metabolism. Novel formulation and drug delivery systems are being investigated to achieve these goals.
One approach involves the development of prodrugs, where the parent compound is chemically modified to improve its absorption, distribution, metabolism, and excretion (ADME) properties. Studies have explored this compound prodrugs, including polymeric forms, with the aim of achieving prolonged biological action and improved bioavailability. researchgate.netakjournals.com While some early polymeric prodrugs did not show improved worm burden reduction compared to this compound, the concept of using prodrug strategies to enhance pharmacokinetic profiles remains an active area of research. nih.gov
Liposomal encapsulation is another delivery system being investigated. Liposome-entrapped this compound (LOXA) has been tested in murine models of S. mansoni infection. LOXA administered subcutaneously demonstrated a significant reduction in worm burden compared to free this compound, particularly when given around the time of infection. Maximum efficacy, with a 97% reduction in parasite number, was observed with subcutaneous LOXA administered one day before infection. scielo.br This suggests that liposomal formulations can influence the delivery and efficacy of this compound.
Research also focuses on understanding the pharmacokinetic/pharmacodynamic (PK/PD) relationship of this compound and its analogs to inform formulation development. Studies have identified an in vitro-in vivo paradox with this compound, where maximal clinical plasma concentrations are significantly lower than the in vitro concentrations required for worm killing. nih.govresearchgate.net Modeling suggests that the parasite's location in the portal vasculature means that portal concentrations are more relevant to efficacy than systemic plasma concentrations. nih.govresearchgate.net This understanding guides the design of formulations aimed at achieving higher and sustained concentrations in the portal circulation.
Improving physicochemical properties like solubility and permeability is also crucial for optimizing oral absorption and achieving favorable portal concentrations. nih.govresearchgate.net Novel this compound analogs are being designed with optimized properties to enhance oral bioavailability and improve in vivo activity. nih.gov
Additionally, research into novel oral formulations, such as floating tablets, has been explored to potentially prolong gastric residence time and improve drug absorption.
Q & A
Q. What analytical methodologies are recommended for quantifying oxamniquine in pharmaceutical formulations and biological matrices?
A validated spectrofluorimetric method using derivatization with 1-dimethylaminonaphthalene-5-sulphonyl chloride (dansyl chloride) is widely employed. This method achieves linearity at 0.02–0.2 µg ml⁻¹, with a detection limit of 0.007 µg ml⁻¹ and quantitation limit of 0.02 µg ml⁻¹ . Key parameters include reaction at pH 10 (sodium carbonate buffer), excitation/emission wavelengths of 335/445 nm, and robustness against minor variations in reagent volume (±0.1 ml) or reaction time (±5 min) . Cross-validation with HPLC or spectrophotometric methods ensures accuracy in dosage forms and spiked plasma (mean recovery: 97.77% ±1.19) .
Q. How does this compound exert its schistosomicidal activity at the molecular level?
this compound is enzymatically activated by sulfotransferases in Schistosoma mansoni to form a reactive ester intermediate, which alkylates parasitic DNA, disrupting replication . Resistance arises from loss-of-function mutations in the sulfotransferase gene (SmSULT), as shown via linkage mapping (LOD = 31 on chromosome 6) and RNAi knockdowns . Crystallographic studies confirm drug-enzyme binding interactions, guiding rational derivative design for broader species efficacy .
Q. What are the primary considerations for designing in vitro assays to evaluate this compound efficacy?
Use juvenile (1–5-day-old) and adult (25-day-old) schistosomula to assess stage-specific susceptibility. Statistical analysis via non-parametric tests (e.g., Kruskal-Wallis) is critical, as strain- and sex-dependent responses exist. For example, R1 strain females show absolute resistance post-day 25, while males exhibit partial susceptibility (17.7% reduction vs. 69.2% in LE strain) . Include controls for baseline parasite viability and validate results across multiple biological replicates.
Advanced Research Questions
Q. How can contradictory data on this compound’s efficacy across Schistosoma strains be resolved methodologically?
Contradictions arise from genetic heterogeneity (e.g., SmSULT mutations) and hybridization events (e.g., S. bovis x S. haematobium hybrids). Combine whole-genome sequencing with functional assays (e.g., CRISPR-Cas9 knockouts) to map resistance loci . For hybrids, perform SNP analysis and in vitro drug exposure trials to quantify hybrid-specific susceptibility . Standardize protocols for parasite age, culture conditions, and drug exposure times to minimize variability .
Q. What experimental strategies are recommended to mitigate false positives/negatives in this compound resistance studies?
- False positives: Use isogenic parasite lines to control for genetic background effects. Confirm resistance via enzymatic assays (e.g., sulfotransferase activity with PAPS cofactor) .
- False negatives: Optimize drug concentration ranges using dose-response curves (e.g., IC₅₀ values) and include sensitive strains as internal controls. Apply orthogonal methods like LC-MS to verify metabolite activation .
Q. How can spectrofluorimetric methods be adapted for high-throughput screening of this compound derivatives?
Automate derivatization steps using microplate readers and robotic liquid handlers. Validate derivative-specific fluorescence profiles (e.g., excitation/emission shifts) and cross-reference with LC-MS for structural confirmation. Use spiked plasma pools to assess matrix effects and recovery rates .
Q. What are the implications of this compound’s species-specific activation for cross-species schistosomiasis control?
S. haematobium lacks functional sulfotransferase orthologs, rendering this compound ineffective. Structure-activity relationship (SAR) studies guided by SmSULT crystallography can identify derivatives with broader specificity. For example, modifying the quinoline scaffold or esterification site may enhance binding to divergent sulfotransferases .
Methodological Resources
- Analytical Validation: Follow ICH Q2B guidelines for specificity, linearity, accuracy, and precision .
- Genetic Mapping: Utilize CRISPR-Cas9 and RNAi for functional genomics in schistosomes .
- Data Reporting: Adhere to standards for qualitative research (e.g., SRQR) and include raw data in supplemental files .
Retrosynthesis Analysis
AI-Powered Synthesis Planning: Our tool employs the Template_relevance Pistachio, Template_relevance Bkms_metabolic, Template_relevance Pistachio_ringbreaker, Template_relevance Reaxys, Template_relevance Reaxys_biocatalysis model, leveraging a vast database of chemical reactions to predict feasible synthetic routes.
One-Step Synthesis Focus: Specifically designed for one-step synthesis, it provides concise and direct routes for your target compounds, streamlining the synthesis process.
Accurate Predictions: Utilizing the extensive PISTACHIO, BKMS_METABOLIC, PISTACHIO_RINGBREAKER, REAXYS, REAXYS_BIOCATALYSIS database, our tool offers high-accuracy predictions, reflecting the latest in chemical research and data.
Strategy Settings
| Precursor scoring | Relevance Heuristic |
|---|---|
| Min. plausibility | 0.01 |
| Model | Template_relevance |
| Template Set | Pistachio/Bkms_metabolic/Pistachio_ringbreaker/Reaxys/Reaxys_biocatalysis |
| Top-N result to add to graph | 6 |
Feasible Synthetic Routes
Featured Recommendations
| Most viewed | ||
|---|---|---|
| Most popular with customers |
Disclaimer and Information on In-Vitro Research Products
Please be aware that all articles and product information presented on BenchChem are intended solely for informational purposes. The products available for purchase on BenchChem are specifically designed for in-vitro studies, which are conducted outside of living organisms. In-vitro studies, derived from the Latin term "in glass," involve experiments performed in controlled laboratory settings using cells or tissues. It is important to note that these products are not categorized as medicines or drugs, and they have not received approval from the FDA for the prevention, treatment, or cure of any medical condition, ailment, or disease. We must emphasize that any form of bodily introduction of these products into humans or animals is strictly prohibited by law. It is essential to adhere to these guidelines to ensure compliance with legal and ethical standards in research and experimentation.


