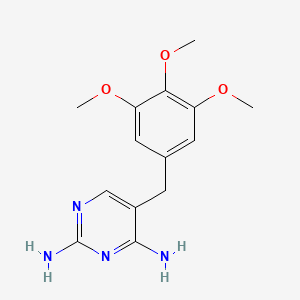
Trimethoprim
Description
Historical Context of Antifolate Agents in Microbiology
The history of antifolate agents in microbiology is rooted in the understanding of folate metabolism, a pathway essential for cell growth and division in many organisms, including bacteria and parasites oup.comnih.govnih.govrcsb.org. The development of these agents began in the 1930s with the discovery of antibacterial activity in synthetic dyes, leading to the identification of sulfanilamide as an active agent in the early 1940s nih.gov. Sulfanilamide was found to mimic para-aminobenzoic acid (pABA), a substrate for dihydropteroate synthase (DHPS), an enzyme critical in the bacterial folate synthesis pathway nih.govnih.gov.
Following the success of sulfonamides, research in the late 1940s by George Hitchings and Gertrude Elion led to the synthesis of substituted 2,4-diaminopyrimidines, which were found to interfere with folate metabolism nih.gov. This research culminated in the discovery of trimethoprim, the first inhibitor of dihydrofolate reductase (DHFR) nih.gov. Initially, antifolates like aminopterin and methotrexate were explored for their anti-leukemic properties in the 1940s, demonstrating their ability to inhibit rapidly dividing cells nih.govnih.govmesotheliomaweb.orgcuni.cz. The success of these agents in treating tumors also paved the way for their application against other rapidly dividing cells, such as bacteria and parasites oup.com. This compound emerged as a highly selective and potent antibacterial agent, particularly against bacterial DHFR enzymes like those found in Staphylococcus aureus and Escherichia coli, with good oral bioavailability nih.gov.
Significance of Dihydrofolate Reductase Inhibition in Antimicrobial Strategies
Dihydrofolate reductase (DHFR) is a vital enzyme for maintaining bacterial growth and replication, making it a significant target for antimicrobial strategies nih.govontosight.airesearchgate.netnih.govoregonstate.edumdpi.com. DHFR catalyzes the reduction of dihydrofolate (DHF) to tetrahydrofolate (THF), a crucial cofactor required for the synthesis of essential building blocks like thymidylate, purines, and certain amino acids (e.g., methionine and histidine) nih.govontosight.ainih.govmdpi.compatsnap.compatsnap.com. These components are indispensable for DNA synthesis and cell division nih.govontosight.aipatsnap.compatsnap.com. By inhibiting DHFR, compounds like this compound prevent the conversion of DHF to THF, leading to a depletion of essential nucleotides and ultimately halting bacterial DNA synthesis, resulting in bacteriostasis ontosight.ainih.govpatsnap.compatsnap.com.
The effectiveness of DHFR inhibitors, including this compound, stems from their ability to selectively target bacterial DHFR over mammalian DHFR nih.govnih.govpatsnap.compatsnap.comnih.gov. This selectivity is attributed to structural differences between the bacterial and mammalian enzymes, allowing for the design of inhibitors that bind more tightly to the bacterial enzyme, thereby minimizing potential side effects and toxicity in the host nih.govpatsnap.compatsnap.com. This targeted mechanism of action makes DHFR inhibitors a cornerstone of antimicrobial therapy, effective against a variety of bacterial species, including Escherichia coli, Staphylococcus aureus, and Streptococcus pneumoniae patsnap.com. The combination of DHFR inhibitors like this compound with dihydropteroate synthase (DHPS) inhibitors (e.g., sulfamethoxazole) creates a synergistic effect by blocking two distinct steps in the bacterial folate biosynthesis pathway, enhancing antibacterial efficacy and broadening the spectrum of activity nih.govnih.govontosight.aimdpi.com.
Scope and Objectives of Contemporary this compound Research
Contemporary research on this compound is primarily driven by the urgent need to combat antibiotic resistance and explore novel applications for this established antimicrobial agent nih.govontosight.airesearchgate.netoregonstate.edupatsnap.comelifesciences.org. A key objective is to understand and overcome resistance mechanisms that bacteria develop against this compound nih.govontosight.aipatsnap.comnih.govelifesciences.orgwikipedia.orgresearchgate.net. Resistance can arise from mutations in the folA gene, which encodes DHFR, leading to reduced affinity for the inhibitor, or through the acquisition of alternative DHFR enzymes that are not inhibited by the drug ontosight.airesearchgate.netnih.govelifesciences.orgwikipedia.org. Studies are focused on discovering compounds that can inhibit both wild-type DHFR and its this compound-resistant variants, potentially delaying the emergence of resistance elifesciences.org.
Another significant area of research involves optimizing the use of this compound, both as a monotherapy and in combination with other agents, to improve efficacy and minimize resistance development ontosight.aipatsnap.commdpi.comcontemporaryobgyn.netasm.orgdoaj.org. This includes investigating optimal dosing strategies and durations of treatment, particularly for common infections like urinary tract infections (UTIs) mdpi.comcontemporaryobgyn.netbmj.comresearchgate.net. Researchers are also exploring novel chemical scaffolds and combination therapies that might overcome existing resistance mechanisms and expand the clinical utility of DHFR inhibitors mdpi.compatsnap.comelifesciences.orgmdpi.com. For instance, studies are examining this compound's potential in treating skin infections, preventing Pneumocystis jirovecii pneumonia (PJP), and even as a treatment for wound healing in specific conditions clinicaltrials.eu. Furthermore, research extends to understanding the environmental fate of this compound, such as its transformation during wastewater treatment, given its presence as a micropollutant nih.gov. The overarching goal is to ensure the continued relevance and effectiveness of this compound in the face of evolving bacterial threats and to uncover new therapeutic avenues.
Data Table: Key Antifolate Agents and Their Primary Targets
| Antifolate Agent | Primary Target Enzyme | Mechanism of Action | Historical Context |
| Sulfanilamide | Dihydropteroate Synthase (DHPS) | Competitively inhibits pABA, blocking folate synthesis. nih.govnih.gov | Discovered in the 1930s, one of the first systemic antibacterials. nih.gov |
| Proguanil | Dihydrofolate Reductase (DHFR) | Inhibits DHFR, preventing THF synthesis. oup.com | First antimalarial antifolate, discovered in 1945. oup.com |
| Pyrimethamine | Dihydrofolate Reductase (DHFR) | Inhibits DHFR, preventing THF synthesis. oup.compatsnap.com | Identified in the late 1940s, widely used antimalarial. oup.com |
| Aminopterin | Dihydrofolate Reductase (DHFR) | Inhibits DHFR, disrupting folate metabolism. nih.govmesotheliomaweb.orgcuni.cz | First clinically useful antifolate (1947) for leukemia. nih.govnih.gov |
| Methotrexate | Dihydrofolate Reductase (DHFR), Thymidylate Synthase (TS), AICAR Transformylase (ATIC) | Potent inhibitor of DHFR, also affects other folate-dependent enzymes; disrupts DNA synthesis. nih.govmesotheliomaweb.orgpatsnap.commdpi.com | Replaced aminopterin in early 1950s due to better therapeutic index. nih.govmesotheliomaweb.orgcuni.cz |
| This compound | Bacterial Dihydrofolate Reductase (DHFR) | Selectively inhibits bacterial DHFR, halting bacterial DNA synthesis. nih.govontosight.ainih.govpatsnap.compatsnap.commdpi.com | Synthesized in the late 1940s, first bacterial DHFR inhibitor. nih.gov |
Data Table: Selectivity of this compound for Bacterial DHFR
| Enzyme Source | This compound Affinity/Selectivity | Impact |
| Bacterial DHFR (e.g., S. aureus, E. coli) | High affinity, potent inhibition nih.govnih.govpatsnap.compatsnap.com | Effectively halts bacterial growth and replication ontosight.aipatsnap.com |
| Mammalian DHFR | Significantly lower affinity nih.govnih.govpatsnap.compatsnap.com | Minimizes host toxicity and side effects patsnap.compatsnap.com |
Properties
IUPAC Name |
5-[(3,4,5-trimethoxyphenyl)methyl]pyrimidine-2,4-diamine | |
|---|---|---|
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI |
InChI=1S/C14H18N4O3/c1-19-10-5-8(6-11(20-2)12(10)21-3)4-9-7-17-14(16)18-13(9)15/h5-7H,4H2,1-3H3,(H4,15,16,17,18) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI Key |
IEDVJHCEMCRBQM-UHFFFAOYSA-N | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Canonical SMILES |
COC1=CC(=CC(=C1OC)OC)CC2=CN=C(N=C2N)N | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Formula |
C14H18N4O3 | |
| Record name | TRIMETHOPRIM | |
| Source | CAMEO Chemicals | |
| URL | https://cameochemicals.noaa.gov/chemical/21180 | |
| Description | CAMEO Chemicals is a chemical database designed for people who are involved in hazardous material incident response and planning. CAMEO Chemicals contains a library with thousands of datasheets containing response-related information and recommendations for hazardous materials that are commonly transported, used, or stored in the United States. CAMEO Chemicals was developed by the National Oceanic and Atmospheric Administration's Office of Response and Restoration in partnership with the Environmental Protection Agency's Office of Emergency Management. | |
| Explanation | CAMEO Chemicals and all other CAMEO products are available at no charge to those organizations and individuals (recipients) responsible for the safe handling of chemicals. However, some of the chemical data itself is subject to the copyright restrictions of the companies or organizations that provided the data. | |
| Record name | trimethoprim | |
| Source | Wikipedia | |
| URL | https://en.wikipedia.org/wiki/Trimethoprim | |
| Description | Chemical information link to Wikipedia. | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
DSSTOX Substance ID |
DTXSID3023712 | |
| Record name | Trimethoprim | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID3023712 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
Molecular Weight |
290.32 g/mol | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Physical Description |
Trimethoprim is an odorless white powder. Bitter taste. (NTP, 1992), Solid | |
| Record name | TRIMETHOPRIM | |
| Source | CAMEO Chemicals | |
| URL | https://cameochemicals.noaa.gov/chemical/21180 | |
| Description | CAMEO Chemicals is a chemical database designed for people who are involved in hazardous material incident response and planning. CAMEO Chemicals contains a library with thousands of datasheets containing response-related information and recommendations for hazardous materials that are commonly transported, used, or stored in the United States. CAMEO Chemicals was developed by the National Oceanic and Atmospheric Administration's Office of Response and Restoration in partnership with the Environmental Protection Agency's Office of Emergency Management. | |
| Explanation | CAMEO Chemicals and all other CAMEO products are available at no charge to those organizations and individuals (recipients) responsible for the safe handling of chemicals. However, some of the chemical data itself is subject to the copyright restrictions of the companies or organizations that provided the data. | |
| Record name | Trimethoprim | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014583 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Solubility |
less than 1 mg/mL at 75 °F (NTP, 1992), ... Very slightly soluble in water and slightly soluble in alcohol., Soluble in N,N-dimethylacetamide (DMAC) at 13.86; benzyl alcohol at 7.29; propylene glycol at 2.57; chloroform at 1.82; methanol at 1.21; ether at 0.003; benzene at 0.002 g/100 ml at 25 °C., In water, 400 mg/l @ 25 °C, 6.15e-01 g/L | |
| Record name | TRIMETHOPRIM | |
| Source | CAMEO Chemicals | |
| URL | https://cameochemicals.noaa.gov/chemical/21180 | |
| Description | CAMEO Chemicals is a chemical database designed for people who are involved in hazardous material incident response and planning. CAMEO Chemicals contains a library with thousands of datasheets containing response-related information and recommendations for hazardous materials that are commonly transported, used, or stored in the United States. CAMEO Chemicals was developed by the National Oceanic and Atmospheric Administration's Office of Response and Restoration in partnership with the Environmental Protection Agency's Office of Emergency Management. | |
| Explanation | CAMEO Chemicals and all other CAMEO products are available at no charge to those organizations and individuals (recipients) responsible for the safe handling of chemicals. However, some of the chemical data itself is subject to the copyright restrictions of the companies or organizations that provided the data. | |
| Record name | Trimethoprim | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00440 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | TRIMETHOPRIM | |
| Source | Hazardous Substances Data Bank (HSDB) | |
| URL | https://pubchem.ncbi.nlm.nih.gov/source/hsdb/6781 | |
| Description | The Hazardous Substances Data Bank (HSDB) is a toxicology database that focuses on the toxicology of potentially hazardous chemicals. It provides information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. The information in HSDB has been assessed by a Scientific Review Panel. | |
| Record name | Trimethoprim | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014583 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Color/Form |
White to cream, crystalline powder | |
CAS No. |
738-70-5 | |
| Record name | TRIMETHOPRIM | |
| Source | CAMEO Chemicals | |
| URL | https://cameochemicals.noaa.gov/chemical/21180 | |
| Description | CAMEO Chemicals is a chemical database designed for people who are involved in hazardous material incident response and planning. CAMEO Chemicals contains a library with thousands of datasheets containing response-related information and recommendations for hazardous materials that are commonly transported, used, or stored in the United States. CAMEO Chemicals was developed by the National Oceanic and Atmospheric Administration's Office of Response and Restoration in partnership with the Environmental Protection Agency's Office of Emergency Management. | |
| Explanation | CAMEO Chemicals and all other CAMEO products are available at no charge to those organizations and individuals (recipients) responsible for the safe handling of chemicals. However, some of the chemical data itself is subject to the copyright restrictions of the companies or organizations that provided the data. | |
| Record name | Trimethoprim | |
| Source | CAS Common Chemistry | |
| URL | https://commonchemistry.cas.org/detail?cas_rn=738-70-5 | |
| Description | CAS Common Chemistry is an open community resource for accessing chemical information. Nearly 500,000 chemical substances from CAS REGISTRY cover areas of community interest, including common and frequently regulated chemicals, and those relevant to high school and undergraduate chemistry classes. This chemical information, curated by our expert scientists, is provided in alignment with our mission as a division of the American Chemical Society. | |
| Explanation | The data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated. | |
| Record name | Trimethoprim [USAN:USP:INN:BAN:JAN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0000738705 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Trimethoprim | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00440 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | trimethoprim | |
| Source | DTP/NCI | |
| URL | https://dtp.cancer.gov/dtpstandard/servlet/dwindex?searchtype=NSC&outputformat=html&searchlist=757370 | |
| Description | The NCI Development Therapeutics Program (DTP) provides services and resources to the academic and private-sector research communities worldwide to facilitate the discovery and development of new cancer therapeutic agents. | |
| Explanation | Unless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source. | |
| Record name | trimethoprim | |
| Source | DTP/NCI | |
| URL | https://dtp.cancer.gov/dtpstandard/servlet/dwindex?searchtype=NSC&outputformat=html&searchlist=106568 | |
| Description | The NCI Development Therapeutics Program (DTP) provides services and resources to the academic and private-sector research communities worldwide to facilitate the discovery and development of new cancer therapeutic agents. | |
| Explanation | Unless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source. | |
| Record name | Trimethoprim | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID3023712 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
| Record name | Trimethoprim | |
| Source | European Chemicals Agency (ECHA) | |
| URL | https://echa.europa.eu/substance-information/-/substanceinfo/100.010.915 | |
| Description | The European Chemicals Agency (ECHA) is an agency of the European Union which is the driving force among regulatory authorities in implementing the EU's groundbreaking chemicals legislation for the benefit of human health and the environment as well as for innovation and competitiveness. | |
| Explanation | Use of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page. | |
| Record name | TRIMETHOPRIM | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/AN164J8Y0X | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
| Record name | TRIMETHOPRIM | |
| Source | Hazardous Substances Data Bank (HSDB) | |
| URL | https://pubchem.ncbi.nlm.nih.gov/source/hsdb/6781 | |
| Description | The Hazardous Substances Data Bank (HSDB) is a toxicology database that focuses on the toxicology of potentially hazardous chemicals. It provides information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. The information in HSDB has been assessed by a Scientific Review Panel. | |
| Record name | Trimethoprim | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014583 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Melting Point |
390 to 397 °F (NTP, 1992), 199-203 °C, 199 - 203 °C | |
| Record name | TRIMETHOPRIM | |
| Source | CAMEO Chemicals | |
| URL | https://cameochemicals.noaa.gov/chemical/21180 | |
| Description | CAMEO Chemicals is a chemical database designed for people who are involved in hazardous material incident response and planning. CAMEO Chemicals contains a library with thousands of datasheets containing response-related information and recommendations for hazardous materials that are commonly transported, used, or stored in the United States. CAMEO Chemicals was developed by the National Oceanic and Atmospheric Administration's Office of Response and Restoration in partnership with the Environmental Protection Agency's Office of Emergency Management. | |
| Explanation | CAMEO Chemicals and all other CAMEO products are available at no charge to those organizations and individuals (recipients) responsible for the safe handling of chemicals. However, some of the chemical data itself is subject to the copyright restrictions of the companies or organizations that provided the data. | |
| Record name | Trimethoprim | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00440 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Trimethoprim | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014583 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Mechanistic Elucidation of Trimethoprim Action
Molecular Basis of Dihydrofolate Reductase (DHFR) Inhibition
The primary molecular target of trimethoprim is dihydrofolate reductase (DHFR), an enzyme vital for bacterial survival. drugbank.compatsnap.com
Competitive Binding to Bacterial DHFR
This compound acts as a competitive inhibitor of bacterial dihydrofolate reductase (DHFR). researchgate.netnih.govvirginia.edu It binds reversibly to the active site of the DHFR enzyme, the same site where the natural substrate, dihydrofolic acid (DHF), would normally bind. open.eduresearchgate.netdrugbank.com By occupying this active site, this compound prevents DHF from binding and being reduced to tetrahydrofolic acid (THF). researchgate.netwikipedia.org This competitive inhibition is a key feature of its mechanism, with this compound effectively "out-competing" the natural substrate. open.edu The binding affinity of this compound for bacterial DHFR is remarkably high, with an inhibition constant (Ki) in the nanomolar range, underscoring the potency of its inhibitory action. numberanalytics.com
Structural Analogy to Dihydrofolic Acid
The efficacy of this compound as a competitive inhibitor stems from its structural similarity to dihydrofolic acid (DHF). open.eduresearchgate.nettandfonline.com Specifically, this compound is a structural analogue of the pteridine portion of dihydrofolic acid. virginia.edulibretexts.org This structural mimicry allows it to fit into the active site of the DHFR enzyme. open.edu The 2,4-diaminopyrimidine moiety of this compound is a key feature that facilitates this interaction. basicmedicalkey.com
Downstream Biochemical Consequences of DHFR Inhibition
The inhibition of DHFR by this compound sets off a cascade of biochemical events that are detrimental to the bacterial cell.
Disruption of Tetrahydrofolate Coenzyme Synthesis
The immediate and primary consequence of DHFR inhibition is the blockade of the conversion of dihydrofolic acid (DHF) to tetrahydrofolic acid (THF). patsnap.comtandfonline.comnih.gov THF is the biologically active form of folic acid and serves as a crucial coenzyme in a variety of metabolic one-carbon transfer reactions. researchgate.nettandfonline.com By preventing the synthesis of THF, this compound effectively depletes the intracellular pool of this essential coenzyme. basicmedicalkey.compatsnap.com This disruption of the folate cycle is the central event that leads to the downstream effects on bacterial metabolism and growth. basicmedicalkey.com
Impact on Purine and Pyrimidine Biosynthesis
The depletion of tetrahydrofolate (THF) has a profound impact on the de novo synthesis of purines and pyrimidines, which are the fundamental building blocks of DNA and RNA. open.edudrugbank.compatsnap.comtandfonline.com THF derivatives are essential for the synthesis of thymidylate, a necessary component of DNA. researchgate.netnih.gov The inhibition of DHFR leads to a reduction in the availability of these THF cofactors, thereby disrupting the production of nucleotides required for DNA replication and RNA synthesis. researchgate.netpatsnap.comnih.gov Studies have shown that treatment with this compound leads to the inhibition of DNA, RNA, and protein synthesis. nih.gov In some bacteria, such as Bacillus subtilis, purine depletion has been identified as a major bottleneck caused by this compound treatment. nih.gov
Effects on Bacterial DNA Synthesis and Cell Proliferation
This compound's primary mechanism of action is the disruption of bacterial DNA synthesis, which subsequently inhibits cell proliferation. wikipedia.orgpatsnap.comnumberanalytics.com This is achieved through the specific inhibition of dihydrofolate reductase (DHFR), a crucial enzyme in the folic acid metabolic pathway. patsnap.comnumberanalytics.comfrontiersin.org
Bacterial cells synthesize their own folic acid, an essential component for the production of nucleotides, the building blocks of DNA. patsnap.com The enzyme DHFR is responsible for the reduction of dihydrofolic acid (DHF) to tetrahydrofolic acid (THF). wikipedia.orgnumberanalytics.com THF is a vital precursor in the synthesis of thymidine, a nucleotide required for DNA replication. wikipedia.orgnumberanalytics.com
This compound acts as a competitive inhibitor of bacterial DHFR, binding to the enzyme's active site with a significantly higher affinity than its natural substrate, DHF. patsnap.comnumberanalytics.com This binding is approximately sixty thousand times stronger for bacterial DHFR compared to human DHFR, which accounts for its selective toxicity against bacteria. wikipedia.org By blocking DHFR, this compound effectively halts the production of THF, leading to a depletion of the cellular pool of this essential co-factor. patsnap.comnumberanalytics.com The absence of THF prevents the synthesis of thymidylate, which in turn arrests DNA replication and, consequently, bacterial growth and cell division. patsnap.comnumberanalytics.com While this compound is considered bacteriostatic when used alone, its combination with sulfamethoxazole, which inhibits an earlier step in the folate pathway, can be bactericidal. patsnap.comdrugbank.com
The inhibition of the folate pathway and the subsequent depletion of essential metabolites can also induce a state known as thymineless death. asm.orgpnas.org This phenomenon is characterized by the stalling of DNA replication forks, leading to DNA damage and ultimately cell death. pnas.orgnih.gov The bactericidal effect of this compound is more pronounced in rich media, where the primary trigger for cell death is accelerated thymine depletion. pnas.orgnih.gov
Investigating Alternative or Ancillary Mechanisms of Action
While the primary mechanism of this compound is the inhibition of DHFR, research has explored other potential ways the drug and its derivatives exert their effects.
DNA-Binding Effects of this compound Analogs
Recent studies have focused on synthesizing this compound analogs with modified structures to investigate additional biological activities. A key modification has been the introduction of an amide bond in place of the methylene bridge found in the parent this compound molecule. mdpi.comnih.gov This alteration was designed to confer DNA-binding properties to the analogs. mdpi.com
Research has shown that while this compound itself does not bind to plasmid DNA, several of its analogs with an amide bond demonstrate significant DNA-binding capabilities. mdpi.comnih.gov For instance, certain analogs exhibited a higher binding affinity to pBR322 plasmid DNA compared to the known DNA-binding agent netropsin. nih.gov The introduction of the amide bond appears to be a crucial factor in this enhanced DNA interaction. nih.gov
Further investigations using various DNA types, including calf thymus and T4 coliphage DNA, have suggested that these analogs may bind within the minor groove of the DNA helix. researchgate.net Molecular docking studies have supported these findings, indicating that the modified structure allows for interactions with DNA. This dual activity of DHFR inhibition and DNA binding presents a promising avenue for developing more potent antimicrobial agents. mdpi.com
| Compound/Analog | Key Structural Modification | DNA Binding Activity | Reference |
| This compound (TMP) | Unmodified | Does not bind to plasmid DNA | mdpi.com |
| TMP Analogs (general) | Introduction of an amide bond | Increased affinity for human DHFR and DNA binding | mdpi.com |
| Analogs 1-6 | Amide bond in place of methylene linker | Compounds 2, 3, and 5 showed higher binding to pBR322 plasmid DNA than netropsin | nih.gov |
| Analogs with Amide Bond | Amide bond | Bind to DNA in AT-rich regions |
Modulation of Cellular Metabolism Beyond Folate Pathway
The inhibition of the folate pathway by this compound has downstream effects on various cellular metabolic processes. The depletion of tetrahydrofolate, a key one-carbon donor, impacts the synthesis of purines, amino acids like methionine and glycine, and S-adenosylmethionine (SAM). researchgate.netnih.gov
Furthermore, research indicates that this compound-induced DNA replication stress can perturb cellular metabolism, leading to the production of reactive oxygen species (ROS) under aerobic conditions. pnas.orgnih.gov Under anaerobic conditions, this stress can result in the production of DNA-damaging byproducts of nitrate respiration. pnas.orgnih.gov This suggests a link between the primary action on the folate pathway and broader metabolic disturbances that contribute to the drug's bactericidal effect. pnas.orgnih.gov
Metabolomic studies have also revealed that this compound can cause pH-dependent effects on amino acid profiles and increase the levels of the osmoprotectant trehalose, indicating a cellular stress response. plos.org These findings highlight that the impact of this compound extends beyond the direct inhibition of folate synthesis, influencing a network of interconnected metabolic pathways. researchgate.netplos.org
Bacterial Resistance to Trimethoprim
Emergence and Spread of Resistance
The emergence and dissemination of trimethoprim resistance are complex processes driven by various genetic and environmental factors.
Horizontal gene transfer (HGT) is a predominant mechanism facilitating the rapid spread of antibiotic resistance genes, including those conferring this compound resistance, among bacterial populations. bioguardlabs.commdpi.comnih.govfrontiersin.org This process involves the transfer of genetic material between bacteria through three main mechanisms:
Conjugation: Considered the most prevalent method for the dissemination of antibiotic resistance genes, conjugation involves direct cell-to-cell contact and the transfer of DNA, typically via plasmids. bioguardlabs.commdpi.comnih.govfrontiersin.org
Transduction: Involves bacteriophages (viruses that infect bacteria) transferring bacterial DNA from one bacterium to another. bioguardlabs.commdpi.comnih.govfrontiersin.org
Transformation: Refers to the uptake of naked extracellular DNA fragments by a recipient bacterium. mdpi.comnih.govfrontiersin.org
Mobile genetic elements (MGEs) such as plasmids, integrons, and transposons play a crucial role in mobilizing and disseminating this compound resistance genes. nih.govfrontiersin.orgmicrobiologyresearch.orgoup.comnih.govuit.nomdpi.com Specifically, dihydrofolate reductase (dfr) genes, which confer resistance to this compound by encoding resistant DHFR enzymes, are frequently found on these MGEs. frontiersin.orgoup.comnih.govuit.no Class 1 and 2 integrons are particularly associated with dfrA genes, often alongside sul genes (conferring sulfamethoxazole resistance), forming "clinical integrons" that contribute to multidrug resistance. frontiersin.orgoup.com The selective pressure exerted by the widespread use of this compound and its combination with sulfamethoxazole (cotrimoxazole) promotes the evolution of increasingly compact gene arrangements carried by these mobile genetic elements, facilitating their movement within and between bacterial genomes. frontiersin.orgnih.gov For instance, studies on uropathogenic Escherichia coli have identified dfrA14 or dfrA1 gene cassettes integrated into conserved sul2-strA-strB clusters on transferable plasmids, ensuring co-resistance. frontiersin.orgnih.gov
Global and regional surveillance systems are critical for monitoring the prevalence and trends of antimicrobial resistance (AMR), including resistance to this compound. The World Health Organization's (WHO) Global Antimicrobial Resistance and Use Surveillance System (GLASS) is a key initiative that collects and reports national data on AMR and antimicrobial use in humans. who.intwho.int
Data from surveillance efforts highlight the significant burden of bacterial AMR, particularly in urinary tract infections (UTIs), where Escherichia coli and Klebsiella pneumoniae are major pathogens. nih.govnih.gov In 2019, this compound/sulfamethoxazole-resistant Escherichia coli alone was attributed to 3,453 deaths globally. nih.gov
Regional surveillance data reveal varying patterns of this compound resistance. For example, a systematic review of antimicrobial resistance among uropathogens in the Asia-Pacific region (APAC) from 2008 to 2020 indicated a very high prevalence of resistance to this compound/sulfamethoxazole, ranging between 33% and 90%. nih.gov The highest prevalence rates were reported from countries such as Bangladesh, India, Sri Lanka, and Indonesia. nih.gov
The following table illustrates the point prevalence estimates of resistance to co-trimoxazole (this compound/sulfamethoxazole) in E. coli and K. pneumoniae in selected Asia-Pacific countries:
| Country | Surveillance Strategy | Co-trimoxazole Resistance (%) |
| Bangladesh | Laboratory-based | 58.0 nih.gov |
| Bhutan | Laboratory-based | 52.9 nih.gov |
| India | Laboratory-based | 64.2 - 73.9 nih.gov |
| Indonesia | Population-based | >50 nih.gov |
Horizontal Gene Transfer
Overcoming Resistance: Research Strategies
To combat the escalating problem of this compound resistance, research efforts are focused on two primary strategies: identifying novel DHFR inhibitors and developing new this compound analogs.
Research is actively pursuing the discovery of new compounds that can effectively inhibit both wild-type bacterial DHFR and its this compound-resistant mutant variants. elifesciences.orgnih.govasm.orgresearchgate.netrsc.orgnih.gov This involves screening large compound databases and employing structure-based design approaches. elifesciences.orgnih.govresearchgate.net
One promising approach involves identifying inhibitors with novel chemical scaffolds. For instance, a compound designated CD15-3 was identified through an integrated computational and experimental approach. elifesciences.org CD15-3 demonstrated inhibition of wild-type E. coli DHFR and its this compound-resistant variants (P21L, A26T, and L28R) with comparable IC50 values (50–75 µM). elifesciences.org Notably, the emergence of resistance to CD15-3 was significantly delayed compared to this compound in in vitro evolution experiments, and whole-genome sequencing of resistant strains showed no mutations in the target folA locus. elifesciences.org
Another class of compounds under development is propargyl-linked antifolates (PLAs). These inhibitors are designed to be potent against a range of DHFR-containing bacteria, including wild-type and this compound-resistant strains of Methicillin-Resistant Staphylococcus aureus (MRSA). asm.orgresearchgate.netrsc.orgnih.gov Studies have shown that PLAs can retain potency against resistant strains, with some compounds exhibiting low mutational frequencies and maintaining effective MIC values. asm.org
Furthermore, ionized non-classical antifolates (INCAs) represent a novel class of DHFR inhibitors developed using a structure-based approach. nih.gov These compounds are engineered to broadly inhibit resistance-conferring DHFR isoforms, including those found in clinical Staphylococcus aureus isolates like DfrG and DfrK, which render this compound and iclaprim ineffective. nih.gov INCAs have demonstrated low nanomolar enzymatic activity and potent cellular activity with human selectivity against clinically relevant this compound-resistant MRSA isolates. nih.gov
Computational methods, such as density-functional calculations, molecular docking, and molecular dynamics simulations, are increasingly utilized to design lead compounds that target atypical plasmid-encoded DHFRs, such as R67 DHFR (Type II DHFR), which bears no structural resemblance to classical DHFRs and confers this compound resistance. mdpi.com
The development of new this compound analogs involves modifying the existing molecular structure to enhance its activity against resistant strains and improve its pharmacodynamic properties without compromising its affinity for bacterial DHFR. mdpi.com
One innovative strategy explores the development of photoresponsive this compound analogs, where the antibacterial activity can be modulated by light. mdpi.com Researchers have reported red-shifted responsive azobenzene photoswitches containing this compound as a core, demonstrating the potential for controlled antibacterial action. mdpi.com
This compound derivatives are also being developed to enhance the drug's ability to suppress DHFR and overcome bacterial resistance. These modifications aim to increase the probability of binding to target proteins and improve treatment efficacy. Examples include:
Halogenated this compound Derivatives: Iodinated this compound (TMP-I) has shown promise as an effective derivative, exhibiting resistance-overcoming properties and synergistic effects with sulfamethoxazole, similar to this compound. Its ease of synthesis from inexpensive starting materials makes it an attractive candidate for further development against MRSA infections.
C7-substituted Analogs: Research is also exploring C7-substituted analogs of this compound, aiming to identify compounds with enhanced affinity and selectivity for bacterial DHFR, thereby circumventing the impact of resistance mutations.
The goal of these analog development efforts is to create robust drugs that are less susceptible to the rapid emergence of resistance, ensuring continued therapeutic options for bacterial infections.
Pharmacodynamics of Trimethoprim
Pharmacodynamic Indices and Their Correlation with Efficacy
Key pharmacodynamic parameters have been identified to correlate with the therapeutic efficacy of antibiotics in both in vitro and in vivo animal infection models tandfonline.com. For trimethoprim, especially in combination with sulfamethoxazole (TMP/SMX), the relationship between drug exposure and bacterial killing can exhibit characteristics of both time-dependent and concentration-dependent activity, depending on the pathogen and specific conditions asm.orgumich.edu.
The free area under the concentration-time curve to minimum inhibitory concentration ratio (fAUC/MIC) is a pharmacodynamic index that combines both the duration and extent of drug exposure researchgate.net. It is considered a predictive pharmacokinetic/pharmacodynamic (PK/PD) index for potentiated sulfonamides like TMP/SMX researchgate.net.
Studies have shown that for Escherichia coli, fAUC/MIC ratios of 40.7, 59.5, and 86.3 were required for stasis, 1-log₁₀, and 2-log₁₀ CFU reductions, respectively, when treated with TMP/SMX nih.govresearchgate.net. In contrast, for Stenotrophomonas maltophilia, no quantifiable fAUC/MIC thresholds for stasis or CFU reductions were observed, indicating a lack of consistent activity against this pathogen even at varying MICs nih.govresearchgate.netresearchgate.net.
For this compound as a component of TMP/SMX, stasis against Stenotrophomonas maltophilia in an in vitro chemostat model was achieved at an fAUC/MIC of 67.4 researchgate.netnih.govoup.com. However, clinically meaningful exposures required to achieve 1-log₁₀ CFU reductions were not quantifiable against this pathogen researchgate.netnih.gov.
In a neutropenic murine thigh infection model using TMP/SMX against Staphylococcus aureus, the free AUC/MIC of sulfamethoxazole (SMX) was found to be a better predictor of in vivo antimicrobial activity than Cmax/MIC, with an R² value of 0.69 nih.gov. For this compound, the 24-hour fAUC was 11.3 μg·h/ml (range 7.2–15.2), and the fAUC/MIC ratio for E. coli was 23 (range 16.4–29.2) researchgate.net.
Table 1: fAUC/MIC Ratios for TMP/SMX Against E. coli and S. maltophilia
| Endpoint | E. coli (fAUC/MIC) nih.govresearchgate.net | S. maltophilia (fAUC/MIC) researchgate.netnih.govoup.com |
| Stasis | 40.7 | 67.4 |
| 1-log₁₀ CFU Reduction | 59.5 | Not Quantifiable |
| 2-log₁₀ CFU Reduction | 86.3 | Not Quantifiable |
The free maximum concentration to minimum inhibitory concentration ratio (fCmax/MIC) is another pharmacodynamic index used to predict antimicrobial efficacy redemc.net. For TMP/SMX, fCmax/MIC has been identified as an equivalent pharmacodynamic driver to fAUC/MIC in some studies researchgate.netnih.govoup.com. In an in vitro chemostat model, both fAUC/MIC and fCmax/MIC were identified as pharmacodynamic drivers for TMP/SMX against Stenotrophomonas maltophilia, with stasis achieved at an fAUC/MIC of 67.4 and an fCmax/MIC that correlated similarly researchgate.netnih.govoup.com.
In a study examining TMP/SMX against glycopeptide-intermediate Staphylococcus aureus (GISA), at simulated peak serum concentrations (Cmax), TMP/SMX was bactericidal against two of three strains, suggesting a concentration-dependent antibacterial activity umich.edu. High drug concentration:MIC ratios (9.8–39.2 x MIC) resulted in complete eradication of the organism in two of three strains and a greater than 2-log CFU/ml reduction in the third umich.edu.
The time that the free drug concentration remains above the minimum inhibitory concentration (%T > MIC) is a pharmacodynamic parameter often correlated with the efficacy of time-dependent antibiotics mdpi.comtandfonline.com. While TMP/SMX can exhibit concentration-dependent killing, some data suggest that for antibiotics with a time-dependent action, such as TMP/SMX, improvements in clinical outcomes plateau when the time period in which the concentration exceeds the MIC₉₀ is more than 60% asm.org.
However, in a study using a neutropenic murine thigh infection model for TMP/SMX against Staphylococcus aureus, TMP/SMX did not show antimicrobial activity when the free %T > MIC was less than 100% nih.gov. This suggests that for optimal activity, a high percentage of time above MIC might be necessary for TMP/SMX against certain pathogens in vivo nih.gov.
fCmax/MIC (Free Maximum Concentration to Minimum Inhibitory Concentration Ratio)
In Vitro Pharmacodynamic Models
In vitro pharmacodynamic models are valuable tools for simulating in vivo human pharmacokinetic profiles and assessing the efficacy of antibiotics, including the potential for drug resistance development plos.org. These models allow for controlled study of the drug-pathogen interaction.
Chemostat models are a type of in vitro pharmacodynamic model used to study bacterial growth and kill curves under controlled conditions, simulating drug concentrations over time nih.govresearchgate.netoup.complos.org.
In studies using an in vitro chemostat model, TMP/SMX demonstrated significantly greater killing for Escherichia coli than for Stenotrophomonas maltophilia at various MICs nih.govresearchgate.netresearchgate.net. For E. coli, mean changes in 24-hour CFU ranged from -4.49 to +1.83 log₁₀ CFU depending on the MIC nih.govresearchgate.net. However, against S. maltophilia, TMP/SMX showed no stasis or CFU reductions regardless of the MIC, and no pharmacodynamic thresholds were quantifiable nih.govresearchgate.netresearchgate.net. This suggests limited activity of TMP/SMX monotherapy against susceptible S. maltophilia even at higher doses researchgate.netnih.govoup.com.
Chemostat models have also been used to investigate the efficacy of TMP/SMX in combination with other antibiotics, such as rifampicin, against methicillin-resistant Staphylococcus aureus (MRSA) plos.org. These models can demonstrate the usefulness of combination therapy, particularly against resistant strains, where regrowth observed with monotherapy might be inhibited by the combination plos.org.
In Vivo Pharmacodynamic Models
In vivo pharmacodynamic models, often utilizing animal infection models, are crucial for evaluating the efficacy of antibiotics in a living system, providing insights that complement in vitro findings.
A neutropenic murine thigh infection model has been used to examine the pharmacokinetics/pharmacodynamics of TMP/SMX against Staphylococcus aureus researchgate.netnih.gov. In this model, the antimicrobial efficacy was calculated as the change in bacterial density after 24 hours of treatment nih.gov. The free AUC/MIC and free %T > MIC of sulfamethoxazole were found to correlate better with in vivo antimicrobial activity than Cmax/MIC nih.gov. Specifically, the free AUC/MIC had an R² of 0.69, and free %T > MIC had an R² of 0.71, compared to Cmax/MIC with an R² of 0.53 nih.gov. The study also indicated that distributed doses of TMP/SMX showed higher antimicrobial activity than a single dosage nih.gov. However, antimicrobial activity was not observed when the free %T > MIC was below 100% nih.gov.
For this compound, typical values for apparent volume of distribution, clearance, and terminal half-life were 1.99 L/kg, 0.33 L/h·kg, and 4.2 hours, respectively, in a non-linear mixed effects model fitted to experimental data researchgate.net. The 24-hour fAUC for this compound was 11.3 μg·h/ml researchgate.net.
Table 2: Correlation of PK/PD Indices with in vivo Antimicrobial Activity of TMP/SMX Against S. aureus (Murine Thigh Infection Model) nih.gov
| PK/PD Index | R² Value |
| fAUC/MIC | 0.69 |
| fCmax/MIC | 0.53 |
| %T > MIC | 0.71 |
Neutropenic Murine Thigh Infection Models
Neutropenic murine thigh infection models are frequently employed to elucidate the pharmacokinetic/pharmacodynamic (PK/PD) relationships of antimicrobial agents like this compound, often in combination with sulfamethoxazole (TMP/SMX). These models help define the exposure-response relationship and identify the PK/PD index that best correlates with in vivo antimicrobial efficacy.
A study published in 2020 investigated the PK/PD of TMP/SMX against Staphylococcus aureus using a neutropenic murine thigh infection model. Five S. aureus isolates with TMP/SMX minimum inhibitory concentrations (MICs) ranging from 0.032 to 64 μg/mL were tested. The study found that for sulfamethoxazole, the free area under the blood concentration-time curve to MIC ratio (free AUC/MIC) and the free percentage of time that the concentration remains above the MIC (free %T > MIC) were better correlated with the in vivo antimicrobial activity than the peak concentration to MIC ratio (Cmax/MIC). Specifically, the free AUC/MIC exhibited an R² value of 0.69, while the free %T > MIC showed an R² value of 0.71, both indicating a stronger correlation compared to the free Cmax/MIC with an R² of 0.53. The study also observed that distributed doses of TMP/SMX (2-3 times per day) demonstrated higher antimicrobial activity than a single daily dosage. Furthermore, TMP/SMX did not exhibit antimicrobial activity when the free %T > MIC was below 100%.
Another study, focusing on iclaprim (a pyrimidine structurally related to this compound), also utilized the neutropenic murine thigh infection model to define its PK/PD index. For Staphylococcus aureus ATCC 29213, the 24-hour AUC/MIC index was most closely linked to efficacy, with an R² of 0.65. For Staphylococcus pneumoniae ATCC 10813, both the 24-hour AUC/MIC and the percentage of time that drug concentrations remained above the MIC (%T>MIC) were strongly associated with the effect, both yielding an R² of 0.86.
The following table summarizes key PK/PD correlation findings from studies utilizing neutropenic murine thigh infection models:
| PK/PD Parameter | Pathogen | R² Value (Correlation with Efficacy) | Source |
| Free AUC/MIC (SMX) | Staphylococcus aureus | 0.69 | |
| Free %T > MIC (SMX) | Staphylococcus aureus | 0.71 | |
| Free Cmax/MIC (SMX) | Staphylococcus aureus | 0.53 | |
| 24-h AUC/MIC (Iclaprim) | Staphylococcus aureus ATCC 29213 | 0.65 | |
| 24-h AUC/MIC (Iclaprim) | Staphylococcus pneumoniae ATCC 10813 | 0.86 | |
| %T > MIC (Iclaprim) | Staphylococcus pneumoniae ATCC 10813 | 0.86 |
Challenges in Translational Pharmacodynamic Studies
Translational pharmacodynamic studies, particularly those involving animal models for dihydrofolate reductase inhibitors like this compound, face specific challenges. A notable limitation arises from the significant difference in plasma thymidine concentrations between rodents and humans. Rodents possess considerably higher levels of thymidine in their plasma compared to humans.
This disparity can impact the translatability of in vivo animal infection models, as high exogenous thymidine levels can potentially bypass or reduce the inhibitory effect of DHFR inhibitors on bacterial DNA synthesis, thereby influencing the observed antimicrobial efficacy in these models. Consequently, data derived from such animal models for this compound and similar antifolate agents may require careful interpretation and further consideration when extrapolating findings to human clinical settings. This highlights the ongoing need for additional research to fully define the pharmacodynamic targets and ensure optimal dosing strategies for these pathogens.
Molecular Interactions and Structure-activity Relationships of Trimethoprim
Structure-Activity Relationship (SAR) Studies
Trimethoprim's core structure, 2,4-diamino-5-(3,4,5-trimethoxybenzyl) pyrimidine, is fundamental to its inhibitory activity. mdpi.comnih.govmdpi.com The 2,4-diaminopyrimidine moiety is crucial for forming hydrogen bonds with key residues in the DHFR active site. basicmedicalkey.com For instance, in bacterial DHFR, this moiety interacts with residues such as Asp27, Ile5, and Phe92. In human DHFR, important residues for ligand binding include Ile-7, Glu-30, Phe-31, Phe-34, Leu-67, Arg-70, and Val-115. mdpi.com
The trimethoxybenzyl group also plays a significant role, contributing to the selective binding of this compound to various DHFR enzymes. basicmedicalkey.com The main moieties involved in the interaction between TMP and the receptor are the methoxy groups, which act as hydrogen bond acceptors, and the amine and peptide groups, which serve as hydrogen bond donors. mdpi.com Additionally, the aromatic rings of this compound are often involved in π-π stacking interactions within the enzyme's active site. mdpi.com
Structural modifications to the this compound scaffold can significantly impact its potency and selectivity. The introduction of an amide bond into this compound analogs, for example, has been shown to increase their affinity for human DHFR compared to the unmodified compound. mdpi.comnih.govmdpi.com This modification can also lead to stronger inhibitory properties against human DHFR. mdpi.com
Replacing the 4-methoxy group with benzyloxy or phenylethanone groups can improve interactions within the DHFR binding site, thereby increasing activity. However, the effect of increasing the size of the aliphatic chain within TMP analogs is not always straightforward and requires further investigation. mdpi.comnih.gov
For bacterial DHFR, 2,4-diamino-5-(3',4',5'-trisubstituted benzyl)pyrimidines, particularly 3',5'-dialkoxy-4'-trisubstituted analogs, have demonstrated high antibacterial activity and selectivity. sci-hub.se While some 3',5'-dialkyl derivatives show less selectivity than this compound, they can still be at least 100-fold less active against mammalian DHFR than against microbial enzymes. sci-hub.se The selectivity for bacterial DHFRs can be dependent on the nature of the 4'-substituent, with amino analogs showing high selectivity. sci-hub.se Propargyl-linked inhibitors, for instance, have shown significantly increased potency, with some compounds exhibiting 3500-fold greater potency than this compound against human DHFR. mdpi.com
Modifying the pyrimidine ring, such as replacing it with a benzene ring, can slightly reduce affinity but derivatives may still interact with key residues like Glu-30 and Phe-34. mdpi.comnih.gov Inserting an amide bond in place of the methylene bridge can also allow for binding to the minor groove of DNA, indicating a dual activity. mdpi.comnih.gov
Rational drug design approaches are employed to enhance the biological activity and safety profile of this compound derivatives. this compound serves as a valuable model compound for the development of new DHFR inhibitors. mdpi.comnih.gov Strategies include incorporating structural elements like amide bonds, which are typical of DNA-binding ligands such as netropsin, into TMP analogs. mdpi.comnih.gov This can lead to compounds with dual activity, acting as both DHFR inhibitors and DNA-binding agents. mdpi.comnih.gov
The design process often involves optimizing hydrophobic and hydrophilic interactions within the DHFR active site. mdpi.com For example, molecular models of E. coli DHFR have been used to design analogs incorporating 3'-carboxyalkoxy moieties to generate ionic interactions with positively charged active-site residues like Arg-57, leading to exceptionally high affinities. sci-hub.se The ultimate goal of rational design is to develop compounds that can broadly inhibit both wild-type and resistant DHFR isoforms, addressing the challenge of antibiotic resistance. nih.gov
Impact of Substitutions on Potency and Selectivity
Computational Modeling and Simulation
Computational methods, including molecular docking and molecular dynamics simulations, are indispensable tools for elucidating the molecular interactions of this compound and its analogs with DHFR. mdpi.comnih.gov
Molecular docking studies are used to explore the potential binding modes of ligands within the DHFR active site and to confirm their affinity for the enzyme. nih.govmdpi.com These studies provide insights into the binding energies and specific interactions formed between this compound or its analogs and DHFR. mdpi.comnih.govmdpi.com
For instance, molecular docking results show that the introduction of an amide bond into TMP analogs generally increases their affinity for human DHFR compared to unmodified TMP. mdpi.comnih.govmdpi.comnih.gov Methotrexate (MTX), a potent DHFR inhibitor, typically exhibits a lower binding energy (e.g., -9.5 kcal/mol) compared to this compound and its analogs, reflecting its high inhibitory activity. mdpi.comnih.govmdpi.com
Key amino acid residues frequently observed to interact with this compound and its derivatives in DHFR include Glu-30, Phe-34, Ile-7, Ala-9, Ser-59, Thr-56, Tyr-121, Asp-145, Thr-146, Arg-70, and Val-115. mdpi.comnih.govmdpi.commdpi.comnih.gov These interactions often involve hydrogen bonds and π-π stacking interactions. mdpi.comnih.govmdpi.comnih.gov For example, compound 2, a TMP analog with an amide bond, showed a binding energy of -8.3 kcal/mol and formed five hydrogen bonds with Ala-9, Thr-56, Tyr-121, Asp-145, and Thr-146. nih.gov Molecular docking studies have also revealed that increasing the number of methylene groups in the chain linking two aromatic rings in TMP analogs can cause an inversion in the binding mode. nih.gov
Table 1: Binding Energies of Selected DHFR Inhibitors and Analogs
| Compound | Binding Energy (kcal/mol) | Key Interactions (Example) | Source |
| Methotrexate (MTX) | -9.5 | Ile-7, Glu-30, Gln-35, Asn-64, Arg-70, Val-115 (H-bonds); Phe-34 (π-π) | mdpi.com |
| This compound (TMP) | Higher than analogs (e.g., -7.0 for bacterial DHFR) | 2,4-diaminopyrimidine with Asp27, Ile5, Phe92; trimethoxybenzyl group | nih.govscispace.com |
| TMP Analog 2 (amide bond) | -8.3 | Ala-9, Thr-56, Tyr-121, Asp-145, Thr-146 (H-bonds) | nih.gov |
| TMP Analog IV (benzene ring) | -8.0 | Ser-59, Tyr-121, Thr-146, Glu-30 (H-bonds); Phe-34 (π-π) | nih.gov |
| TMP Analog V (pyridine ring) | -8.0 | Ser-59, Tyr-121, Thr-146, Ala-9 (H-bonds); Phe-34 (π-π) | nih.gov |
| TMP Analog JW2 | -7.7 | Gly-117 (H-bond) | mdpi.com |
| TMP Analog JW8 | -7.8 | Asn-64, Arg-70 (H-bond) | mdpi.com |
Molecular dynamics (MD) simulations are employed to provide a dynamic view of protein-ligand interactions, revealing structural changes, binding stability, and conformational dynamics over time. mdpi.comoup.comrsc.orgmorressier.comresearchgate.netresearchgate.net These simulations analyze parameters such as Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Solvent Accessible Surface Area (SASA), and Radius of Gyration (Rg). mdpi.comresearchgate.net
MD simulations have shown that DHFR inhibitors can stabilize the enzyme, with some analogs leading to lower RMSD values compared to the apo-protein, indicating a stabilizing effect. mdpi.commdpi.com For instance, a derivative (compound 13) was found to have a significant impact on protein stabilization, with RMSD oscillating around 1.1 Å over a 20 ns simulation. mdpi.com
The binding of drugs like this compound and methotrexate induces conformational changes in DHFR, particularly affecting flexible regions such as the Met20 loop. rsc.orgnih.gov These changes can decouple global motion and perturb fast motions at distal regions of DHFR, establishing a dynamic link between the substrate binding site and catalytic residues. nih.gov MD simulations are also crucial for understanding how mutations in DHFR confer resistance to this compound by inducing distinct structural changes, which can be exploited for rational drug design. oup.comrsc.org For example, the L28R mutation in E. coli DHFR, which confers this compound resistance, leads to increased substrate affinity due to newly formed hydrogen bonds with the aminobenzoyl glutamate tail of DHF. rsc.org
MD studies can further elucidate the role of water molecules at the binding interface, showing that negative entropy associated with this compound binding in wild-type DHFR can be due to water organization. rsc.org Simulations have also been used to study the dynamics of atypical DHFRs, such as the R67 DHFR, which confers resistance to this compound and is structurally unrelated to canonical DHFRs. tennessee.edunih.gov
Table 2: Molecular Dynamics Simulation Parameters for this compound and Analogs
| Parameter | Observation for this compound/Analogs | Significance | Source |
| RMSD | Lowered compared to apo-protein; oscillation around 1.1 Å for stabilizing analogs | Indicates protein stabilization upon ligand binding | mdpi.commdpi.com |
| RMSF | Increased in binding site residues (e.g., tubulin-TMP complex) | Suggests increased flexibility in binding region | researchgate.net |
| Rg | Increased within acceptable limits (e.g., tubulin-TMP complex) | Implies protein unfolding to accommodate ligand binding | researchgate.net |
| Conformational Changes | Decoupling of global motion; perturbation of fast motions; Met20 loop dynamics | Reveals dynamic effects of drug binding and allosteric modulation | rsc.orgnih.gov |
| Hydrogen Bonds | Formation of new H-bonds (e.g., L28R mutant with DHF); stability over simulation | Crucial for binding affinity and understanding resistance mechanisms | rsc.orgresearchgate.net |
Free Energy Perturbation and MM-PBSA Calculations
Free Energy Perturbation (FEP) and Molecular Mechanics/Poisson-Boltzmann Surface Area (MM-PBSA) are computational methods used to estimate the binding free energy of ligands to proteins. These methods are based on classical molecular dynamics (MD) simulations and aim to provide a robust and accurate evaluation of free energy changes in biomolecular systems nih.govspringernature.com.
FEP involves transforming disappearing atoms into dummy atoms using a single-topology approach, with electrostatic and van der Waals interactions transformed simultaneously via soft-core potentials acs.org. MM-PBSA, and its variant MM-GBSA (Molecular Mechanics/Generalized Born Surface Area), approximate binding free energies by integrating molecular mechanics and solvation effects, thereby enabling precise evaluation of ligand binding affinity nih.govresearchgate.net. These approaches are computationally more expedient than traditional statistical perturbation calculations and have shown versatility in various ligand-protein interactions nih.gov.
While both FEP and MM-PBSA are valuable, the accuracy and reproducibility of results can vary. For instance, MM-PBSA calculations for the same molecular system can show variations exceeding 10 kcal/mol in smaller molecule-protein complexes, and up to 43 kcal/mol for larger or more flexible ligands nih.gov. Similarly, FEP+ calculations have shown variations up to 3.9 kcal/mol from independent simulations, indicating the importance of ensemble averaging for reliable results nih.gov.
3D-Quantitative Structure-Activity Relationship (3D-QSAR)
Three-dimensional quantitative structure-activity relationship (3D-QSAR) studies are computational methods that examine the connection between a molecule's three-dimensional structure and its biological activity scribd.commdpi.com. These analyses establish mathematical relationships between biological action and various structural descriptors, helping to understand how changes in molecular structure affect biological activity scribd.commdpi.com.
For this compound and its analogues, 3D-QSAR studies have been conducted to understand the structural features influencing their inhibitory potency against enzymes like Escherichia coli dihydrofolate reductase (DHFR) nih.gov. Methods such as Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA) are frequently employed in 3D-QSAR to construct predictive models nih.govmdpi.com. These methods model the influence of molecular shape on steric (Lennard-Jones) and electrostatic (Coulomb) effects involved in non-covalent ligand-receptor interactions mdpi.com.
Analysis of contour maps generated from CoMFA and CoMSIA can reveal specific structural modifications that enhance potency. For instance, studies on 2,4-diamino-5-(substituted-benzyl)pyrimidine derivatives (this compound analogues) have shown that substituting electronegative groups at the first and second positions, along with an electropositive group at the third position of the R2 substituent, significantly increases the derivatives' potency nih.gov. This provides valuable insights for designing new this compound analogues with improved affinity nih.gov.
Interactions with Biological Macromolecules (Excluding Target Enzyme)
Beyond its primary target enzyme, dihydrofolate reductase, this compound also interacts with other biological macromolecules, notably human serum albumin (HSA). These interactions are crucial for understanding the drug's transport and distribution within the body.
Interaction with Human Serum Albumin (HSA)
The interaction between this compound (TMP) and human serum albumin (HSA) has been extensively studied using various spectroscopic and molecular modeling techniques researchgate.nettandfonline.comtandfonline.comnih.gov. HSA is the most abundant protein in human blood plasma and plays a vital role in transporting a wide array of endogenous and exogenous compounds due to its numerous binding sites and conformational flexibility tandfonline.comrsc.org.
Experimental results consistently show that this compound quenches the intrinsic fluorescence of HSA, indicating an interaction between the two molecules researchgate.nettandfonline.comtandfonline.comnih.gov. This quenching typically occurs via a static quenching mechanism, implying the formation of a stable ground-state complex between TMP and HSA rather than dynamic collision researchgate.nettandfonline.comnih.govsci-hub.seerpublications.com. The stability of this complex tends to decrease with increasing temperature sci-hub.se.
HSA possesses primary drug-binding regions, often referred to as Sudlow's site I and site II, located in the hydrophobic cavities of subdomains II-A and III-A, respectively rsc.org. Studies have identified that this compound primarily binds to a single site within subdomain IIA of HSA researchgate.nettandfonline.comtandfonline.comnih.govresearchgate.net. Displacement experiments have further verified this specific binding location researchgate.nettandfonline.com. Molecular docking studies corroborate these findings, revealing that TMP effectively binds to the subdomain IIA (site I) of HSA researchgate.net.
The interaction between this compound and HSA is a spontaneous process, characterized by a negative change in Gibbs free energy (ΔG) tandfonline.com. Thermodynamic parameters derived from interaction studies provide insights into the nature of the binding forces. For the TMP-HSA interaction, negative values for both enthalpy change (ΔH) and entropy change (ΔS) typically indicate that hydrogen bonds and van der Waals forces play a major role in stabilizing the complex researchgate.nettandfonline.comtandfonline.comitmedicalteam.pl. Some studies also report hydrophobic interactions contributing to the complex formation tandfonline.comresearchgate.net.
Specific interactions identified through molecular modeling include hydrogen bonds formed between N3 and N4 atoms of TMP and amino acid residues such as Cys 245 and Gly 248 of HSA researchgate.nettandfonline.comtandfonline.com. Hydrophobic interactions involve various carbon atoms of TMP (e.g., C1, C4, C14) with HSA residues like Cys 200 and His 242 researchgate.nettandfonline.comtandfonline.com. Other forces involving oxygen and carbon atoms of TMP with residues like Gln196 and Arg 257 have also been observed researchgate.nettandfonline.comtandfonline.com.
The distance between the this compound (acceptor) and HSA (donor) has been estimated to be approximately 1.67 nm based on Förster's non-radiative energy transfer theory, which is within the range for energy transfer to occur researchgate.nettandfonline.comtandfonline.comnih.gov.
The binding of this compound to HSA induces conformational changes in the protein researchgate.nettandfonline.comtandfonline.comnih.goverpublications.comresearchgate.net. Synchronous fluorescence spectra indicate that TMP can alter the conformation of HSA, leading to an increased polarity around the tryptophan (Trp) residues researchgate.nettandfonline.comtandfonline.com. Tryptophan 214, located in subdomain IIA, is a key fluorescent residue whose microenvironment is affected itmedicalteam.pl.
Further evidence from Fourier-transform infrared (FT-IR) spectroscopy shows a reduction in the α-helix content of HSA upon TMP binding. For instance, the α-helix content was observed to decrease from 51.4% to 46.7%, while the random coil content increased by approximately 5.2% researchgate.nettandfonline.comtandfonline.com. This suggests that the secondary structure of HSA becomes looser or undergoes a partial unfolding as a consequence of TMP binding tandfonline.complos.org. Circular dichroism (CD) spectroscopy also supports that TMP changes the secondary structures of HSA nih.gov. Molecular dynamics simulations further confirm these structural modifications in the second and third structures of HSA upon TMP binding researchgate.net.
Thermodynamic Parameters and Interaction Forces (Hydrogen Bonds, Van der Waals, Hydrophobic)
DNA-Binding Capacity of this compound Analogs
While this compound itself does not typically bind to plasmid DNA or displace ethidium bromide from DNA complexes wikipedia.org, its analogs, particularly those with structural modifications, demonstrate significant DNA-binding capacity. The ethidium displacement test has been widely used to confirm this property in various this compound derivatives wikipedia.orgwikipedia.orgfishersci.finih.govnih.govmims.com. Some this compound analogs have exhibited higher binding affinity to pBR322 plasmid DNA compared to netropsin, a known DNA-binding agent nih.gov.
The association constants for this compound analogs have been determined across different DNA types, including calf thymus DNA, T4 coliphage DNA, poly (dA-dT)2, and poly (dG-dC)2 wikipedia.orgfishersci.finih.govnih.govmims.com. These studies provide quantitative insights into the strength and nature of the drug-DNA interactions. For example, compounds 1–6 showed association constants in the range of 2.4–5.9 × 10^5 M^-1 for calf thymus DNA, indicating moderate interactions nih.gov.
Minor Groove Binding
A notable aspect of the DNA-binding capacity of this compound analogs is their ability to bind within the minor groove of DNA. This has been validated through various tests, particularly with T4 coliphage DNA, where the major groove is sterically hindered by α-glycosylation of 5-(hydroxymethyl)cytidine residues, thereby confirming minor-groove selectivity for compounds exhibiting high binding constants fishersci.finih.govnih.gov.
The incorporation of an amide bond into the structure of TMP analogs is a key modification that facilitates minor groove binding, drawing a parallel to the structural features of netropsin, a classic minor groove binding agent wikipedia.orgfishersci.fi. Netropsin forms non-intercalating bonds with DNA, and its amide NH groups are known to form hydrogen bonds with the C-2 position of thymine and the N-3 position of adenine in the DNA duplex fishersci.fi. This minor groove binding by this compound analogs and their metal complexes can interfere with essential cellular processes such as transcription and replication wikipedia.org.
Table 1: Representative DNA-Binding Affinities of this compound Analogs
| Compound Class/Type | DNA Type | Association Constant (Kapp, M⁻¹) | Binding Mechanism | Reference |
| TMP Analogs (1-6) | Calf Thymus DNA | 2.4 – 5.9 × 10⁵ | Moderate interaction | nih.gov |
| TMP Analogs (1-18) | T4 Coliphage DNA | 0.7 – 6.4 × 10⁵ | Minor Groove Binding | nih.gov |
| TMP Analogs (13-14) | pBR322 Plasmid | 43.85% - 49.89% fluorescence decrease (relative to control) | DNA Binding | nih.gov |
| TMP Analogs (17-18) | pBR322 Plasmid | 41.68% - 42.99% fluorescence decrease (relative to control) | DNA Binding | nih.gov |
| TMP-Metal Complexes | DNA (general) | Not specified | Minor Groove Binding, blocking transcription/replication | wikipedia.org |
Table 2: DHFR Binding Energies of this compound and Select Analogs (Molecular Docking)
| Compound | Binding Energy (kcal/mol) | Key Interacting Residues (hDHFR) | Reference |
| This compound (TMP) | -7.5 (for unmodified TMP) | Glu-30, Phe-34 (general) | wikipedia.orgfishersci.finih.gov |
| TMP Analogs (14, 16) | -8.2 (higher affinity than unmodified TMP) | Glu-30, Tyr-121, Asp-145, Thr-146, Ala-9 | wikipedia.orgnih.gov |
| Benzamide TMP Derivative IV | -8.0 | Ser-59, Tyr-121, Thr-146, Glu-30, Phe-34 | wikipedia.org |
| Benzamide TMP Derivative V | -8.0 | Ser-59, Tyr-121, Thr-146, Ala-9, Phe-34 | wikipedia.org |
| N-(2-aminopyrimidin-5-yl)-2-(3,4,5-trimethoxyphenyl) acetamide | -8.3 (lowest binding energy among tested analogs) | Tyr-121, Asp-145, Thr-146, Ala-9 | wikipedia.org |
Toxicology and Metabolic Disposition Mechanism-focused
Mechanisms of Adverse Reactions Related to Reactive Metabolites
Trimethoprim's bioactivation to reactive metabolites is considered a potential contributing factor to idiosyncratic adverse drug reactions (IADRs) acs.orgnih.govannualreviews.org. These reactive metabolites can covalently bind to proteins, a process hypothesized to initiate immune responses or direct cellular toxicity nih.gov.
One significant mechanism of this compound bioactivation involves its oxidation to a reactive pyrimidine iminoquinone methide intermediate acs.orgku.eduoup.comnih.govscholaris.caacs.org. This intermediate has a protonated molecular ion of m/z 289 and is highly electrophilic nih.govscholaris.ca. The formation of this iminoquinone methide can occur through direct oxidation of this compound or via sequential secondary oxidation of primary, demethylated this compound metabolites ku.edunih.gov.
Glutathione (GSH) plays a crucial role in detoxifying reactive metabolites of this compound acs.orgnih.govnih.govacs.orgnih.gov. In the presence of GSH, human liver microsomes (HLM) produce GSH conjugates, indicating the trapping of these reactive intermediates acs.orgnih.govacs.orgresearchgate.net. Two major GSH adducts, TMPG-1 and TMPG-2, are believed to originate from the iminoquinone methide intermediate acs.org. Additionally, minor GSH adducts (TMPG-3, -4, and -5) have been identified, suggesting the formation of other reactive intermediates like ortho- or para-quinone methides, potentially following multiple O-demethylations acs.orgnih.govacs.org. The scavenging of reactive this compound metabolites by glutathione is considered a protective mechanism in vivo, leading to the formation of urinary mercapturic acids nih.gov.
Cytochrome P450 (CYP) enzymes are extensively involved in the metabolism and bioactivation of this compound ku.edunih.govnih.govdrugbank.commdpi.comnih.gov. Studies using heterologously expressed human P450s have provided insights into the specific roles of various CYP isoforms:
CYP1A2 and CYP3A4: These enzymes are capable of forming the iminoquinone methide intermediate ku.edunih.gov. They also contribute to the production of major GSH adducts (TMPG-1 and TMPG-2) acs.orgnih.govacs.org. CYP3A4 is particularly significant, catalyzing multiple O-demethylation reactions that can lead to quinone methide intermediates ku.edunih.gov.
CYP2D6: This enzyme is primarily responsible for catalyzing both demethylation and the subsequent secondary oxidation step to form quinone-methide species ku.edunih.gov. CYP2D6 also contributes to the formation of minor GSH adducts, such as TMPG-3 acs.orgnih.govacs.org.
Here is a summary of the involvement of key CYP enzymes in this compound metabolism and bioactivation:
| CYP Enzyme | Primary Role in this compound Metabolism/Bioactivation | Associated Metabolites/Intermediates |
| CYP1A2 | Iminoquinone methide formation, major GSH adducts, minor GSH adducts, 3-N-oxide formation acs.orgku.edunih.govnih.govacs.org | Iminoquinone methide, TMPG-1, TMPG-2, TMPG-3, 3-NO-TMP acs.orgku.edunih.govnih.govacs.org |
| CYP3A4 | Iminoquinone methide formation, multiple O-demethylation reactions leading to quinone methide intermediates, major GSH adducts, 4'-demethylation, 1-N-oxide formation, Cα-NAC-TMP, Cα-OH-TMP acs.orgku.edunih.govnih.govacs.orgdrugbank.com | Iminoquinone methide, quinone methide, TMPG-1, TMPG-2, 4'-desmethyl-TMP, 1-NO-TMP, Cα-NAC-TMP, Cα-OH-TMP acs.orgku.edunih.govnih.govacs.orgdrugbank.com |
| CYP2D6 | Demethylation and secondary oxidation to quinone-methide, minor GSH adducts acs.orgku.edunih.govnih.govacs.org | Quinone methide, TMPG-3, TMP-6 (presumptive precursor to TMPG-3) acs.orgku.edunih.govnih.govacs.org |
| CYP2C9 | 3'-demethylation ku.edunih.govdrugbank.com | 3'-desmethyl-TMP ku.edunih.govdrugbank.com |
The formation of reactive metabolites, such as the iminoquinone methide, suggests a mechanism by which this compound could contribute to idiosyncratic toxic responses acs.orgnih.govannualreviews.org. Idiosyncratic adverse drug reactions (IADRs) are unpredictable and can be severe nih.govchildrensmercy.org. The involvement of CYP2D6 in the formation of reactive metabolites is particularly noteworthy because this enzyme displays wide interindividual variability due to genetic polymorphism acs.orgmdpi.com. This genetic variability in CYP2D6 activity may be a factor in the rare adverse drug reactions observed with this compound acs.orgtandfonline.com. Furthermore, the covalent binding of chemically reactive metabolites to cellular proteins can lead to direct organ toxicity or the formation of neoantigens, potentially stimulating an immune response nih.gov.
Role of Cytochrome P450 Enzymes (CYP1A2, CYP3A4, CYP2D6) in Metabolism and Bioactivation
Metabolic Pathways and Metabolite Identification
This compound undergoes oxidative metabolism, primarily in the liver, yielding a variety of stable metabolites acs.orgku.edunih.govdrugbank.comresearchgate.netjst.go.jp.
The major metabolic pathways for this compound include O-demethylation and N-oxidation acs.orgku.edunih.govdrugbank.comresearchgate.netjst.go.jpnih.govtandfonline.com.
O-Demethylation: This pathway involves the oxidative removal of methyl groups from the trimethoxybenzyl moiety of this compound ku.edunih.govdrugbank.comresearchgate.netjst.go.jpnih.govtandfonline.com. The most abundant O-demethylated metabolites identified in humans are 3'-desmethyl-trimethoprim and 4'-desmethyl-trimethoprim ku.edunih.govdrugbank.com. These two metabolites account for approximately 65% and 25% of the total metabolite formation, respectively nih.govdrugbank.com. Further O-demethylation reactions can occur, potentially leading to up to three free hydroxyl groups on the benzylic ring ku.edunih.gov.
N-Oxidation: This pathway involves the oxidation of nitrogen atoms in the pyrimidine ring ku.edunih.govdrugbank.comresearchgate.netjst.go.jpnih.govtandfonline.com. Stable N-oxide metabolites identified include this compound 1-N-oxide (1-NO-TMP) and this compound 3-N-oxide (3-NO-TMP) ku.edunih.govtandfonline.com. N-oxide metabolites generally constitute a minor proportion of the total metabolites, typically less than 5% drugbank.com.
Other identified metabolites include benzylic alcohol (Cα-OH-TMP), formed via oxidation of the methylene bridge, and its N-acetyl cysteine (NAC) adduct (Cα-NAC-TMP), which serves as evidence of this compound bioactivation ku.edunih.gov.
Here is a summary of stable this compound metabolites and their formation pathways:
| Metabolite Name | Pathway of Formation | Relative Abundance (Human) |
| 3'-desmethyl-trimethoprim | O-Demethylation | ~65% of total metabolites nih.govdrugbank.com |
| 4'-desmethyl-trimethoprim | O-Demethylation | ~25% of total metabolites nih.govdrugbank.com |
| This compound 1-N-oxide | N-Oxidation | Minor (<5%) drugbank.com |
| This compound 3-N-oxide | N-Oxidation | Minor (<5%) drugbank.com |
| Cα-OH-trimethoprim | α-Hydroxylation | Minor ku.edunih.govdrugbank.com |
| Cα-NAC-trimethoprim | Adduct formation | Detected in urine nih.govku.edu |
Formation of α-hydroxyTMP
This compound undergoes metabolic transformation primarily in the liver, with approximately 10-20% of an ingested dose being metabolized drugbank.com. Among its various metabolic pathways, α-hydroxylation is a notable biotransformation reaction jst.go.jpnih.govcdhb.health.nz. The addition of a hydroxyl group to this compound results in the formation of α-hydroxythis compound (α-hydroxyTMP), a primary metabolite ontosight.ainih.gov.
Studies involving human liver microsomes (HLM) and NADPH have identified cytochrome P450 3A4 (CYP3A4) as a key enzyme responsible for the bioactivation of this compound, which contributes to the formation of metabolites, including α-hydroxythis compound nih.gov. This hydroxylation can alter the pharmacokinetic profile of the parent compound, influencing its solubility, protein binding, and metabolic stability ontosight.ai. Research indicates that α-hydroxythis compound (Cα-OH-TMP) has the capacity to covalently modify human serum albumin (HSA) in vitro, suggesting its potential involvement in idiosyncratic adverse drug reactions nih.gov. In animal models, such as rats, α-hydroxy-trimethoprim has been detected in urine, accounting for approximately 5% of the excreted radioactivity within an 8-hour period jst.go.jp.
Mechanism of Folate-Related Adverse Effects (e.g., Megaloblastic Anemia, Thrombocytopenia)
This compound's therapeutic action is rooted in its ability to inhibit bacterial dihydrofolate reductase (DHFR) wikipedia.orgelsevier.esmhmedical.compatsnap.commdpi.com. This enzyme is crucial for the conversion of dihydrofolic acid (DHF) to tetrahydrofolic acid (THF), a vital coenzyme essential for the synthesis of purine nucleotides, thymidine (a precursor for DNA replication), and certain amino acids wikipedia.orgpatsnap.commdpi.com. By blocking this critical step, this compound disrupts bacterial DNA synthesis, leading to impaired bacterial cell growth and proliferation wikipedia.orgpatsnap.commdpi.com.
While this compound exhibits a significantly higher inhibitory activity for bacterial DHFR compared to human DHFR (approximately sixty thousand times greater), it can still bind to the human enzyme, albeit with lower affinity wikipedia.orgmhmedical.comenghusen.dk. This interaction with human DHFR can lead to a reduction in serum folate levels and, in some patients, result in folate deficiency enghusen.dk.
Megaloblastic Anemia: Megaloblastic anemia is a type of macrocytic anemia characterized by impaired DNA synthesis during red blood cell production elsevier.eswikipedia.org. When DNA synthesis is compromised, the cell cycle cannot progress effectively, leading to continued cell growth without division, which manifests as macrocytosis and ineffective erythropoiesis elsevier.eswikipedia.org. This compound, by inhibiting human DHFR and consequently depleting tetrahydrofolate, can induce a functional folate deficiency that impairs DNA synthesis in rapidly dividing cells, including hematopoietic precursor cells in the bone marrow elsevier.esacpjournals.orgresearchgate.net. This can result in megaloblastic changes in red blood cells, particularly in individuals with pre-existing low folate stores enghusen.dkacpjournals.org.
Thrombocytopenia: Thrombocytopenia, characterized by a decreased number of circulating platelets, has been reported as a potential adverse effect associated with this compound, particularly when used in combination with sulfamethoxazole (TMP/SMX) wikipedia.orgada.orgada.orgdroracle.ainih.gov. The mechanism underlying this compound-induced thrombocytopenia is largely considered to be an immune-mediated disorder ada.orgdroracle.ainih.gov. It is hypothesized that the drug can induce the formation of autoantibodies that bind to platelet membrane proteins, leading to increased platelet destruction ada.orgnih.gov. This reaction is thought to fall within the "quinine type" immune pathogenesis category, where the presence of the drug enhances the structural affinity between autoantibodies and platelet glycoproteins, thereby facilitating binding and subsequent platelet destruction ada.org. This adverse effect can be severe and potentially life-threatening, and its onset is considered to be dose- and duration-independent in some cases droracle.ainih.gov.
Mechanisms of Other Systemic Effects (e.g., Hyperkalemia, Creatinine Competition)
Beyond its direct effects on folate metabolism, this compound can exert other systemic effects through distinct mechanisms, notably impacting potassium homeostasis and creatinine excretion.
Hyperkalemia: this compound is a recognized cause of hyperkalemia, an elevation in serum potassium levels wikipedia.orggpnotebook.comnih.govunmc.edunih.govtg.org.aukarger.comacpjournals.orgpathway.mdbmj.com. The primary mechanism involves this compound's action on the renal tubules, where it reduces potassium excretion wikipedia.orggpnotebook.comnih.govtg.org.auacpjournals.org. This occurs through the competitive inhibition of epithelial sodium channels (ENaC) located in the distal nephron wikipedia.orgnih.govunmc.edunih.govtg.org.aukarger.comacpjournals.org. This compound's structural similarity to the potassium-sparing diuretic amiloride allows it to mimic amiloride's effect wikipedia.orgnih.govunmc.edunih.govtg.org.aukarger.comacpjournals.org. By blocking ENaC, this compound reduces the reabsorption of sodium, which in turn diminishes the lumen-negative transepithelial voltage, thereby inhibiting potassium secretion into the urine nih.govacpjournals.org. The risk of developing hyperkalemia is heightened in patients receiving higher dosages of this compound, those with underlying renal impairment, or individuals concurrently taking other medications that can impede renal potassium excretion, such as renin-angiotensin system inhibitors or spironolactone gpnotebook.comunmc.edunih.govtg.org.aukarger.compathway.mdbmj.com.
Creatinine Competition: this compound can lead to an apparent increase in serum creatinine levels, which does not necessarily reflect a true decrease in glomerular filtration rate (GFR) wikipedia.orggpnotebook.comtg.org.aubmj.comnih.govcuriousclinicians.comresearchgate.netdroracle.ai. This phenomenon is due to this compound's competitive inhibition of creatinine secretion in the renal tubules wikipedia.orggpnotebook.comtg.org.aubmj.comnih.govcuriousclinicians.comresearchgate.net. Creatinine is not only filtered by the glomeruli but also actively secreted by the proximal tubules via organic cation transporters, specifically OCT2 and MATE1 curiousclinicians.comresearchgate.net. As a weak organic base, this compound competes with creatinine for these same transporters curiousclinicians.com. This competition reduces the tubular secretion of creatinine, causing it to accumulate in the bloodstream and leading to an elevated serum creatinine concentration nih.govcuriousclinicians.comresearchgate.net. This rise in serum creatinine is typically reversible, often appearing within 1-3 days of initiating this compound therapy and resolving within approximately a week after discontinuation nih.govdroracle.ai.
Table 1: Key Metabolic and Toxicological Mechanisms of this compound
| Mechanism Category | Specific Effect | Underlying Mechanism | Relevant Compounds/Enzymes |
| Metabolic Disposition | α-hydroxylation | Hydroxylation of this compound, primarily by CYP3A4. | This compound, α-hydroxythis compound, CYP3A4 |
| Folate-Related Adverse Effects | Megaloblastic Anemia | Inhibition of human dihydrofolate reductase (DHFR), leading to impaired tetrahydrofolate (THF) synthesis and subsequent disruption of DNA synthesis. | This compound, Dihydrofolate Reductase (DHFR), Dihydrofolate (DHF), Tetrahydrofolate (THF) |
| Thrombocytopenia | Immune-mediated platelet destruction, possibly involving drug-dependent autoantibodies. | This compound, Platelet Glycoproteins, Autoantibodies | |
| Other Systemic Effects | Hyperkalemia | Competitive inhibition of epithelial sodium channels (ENaC) in the distal nephron, similar to amiloride, reducing renal potassium excretion. | This compound, Epithelial Sodium Channels (ENaC), Amiloride |
| Creatinine Competition | Competitive inhibition of tubular secretion of creatinine via organic cation transporters (OCT2, MATE1). | This compound, Creatinine, Organic Cation Transporter 2 (OCT2), Multidrug and Toxin Extrusion Protein 1 (MATE1) |
Drug Interactions and Synergism
Synergistic Interactions with Sulfonamides (e.g., Sulfamethoxazole)
The combination of trimethoprim with sulfonamides, such as sulfamethoxazole, is a well-established example of antimicrobial synergism, often referred to as co-trimoxazole. This combination is widely used to treat various bacterial and some fungal infections. nih.govjove.com
The synergistic effect between this compound and sulfamethoxazole stems from their ability to inhibit successive steps in the bacterial tetrahydrofolate (THF) synthesis pathway, which is crucial for bacterial growth and replication. nih.govwikipedia.orgdrugbank.comaap.org
Sulfamethoxazole's Action : Sulfamethoxazole, a sulfonamide, acts as a competitive inhibitor of dihydropteroate synthase. This enzyme is responsible for converting para-aminobenzoic acid (PABA) to dihydrofolic acid (DHF), an early precursor in the folate synthesis pathway. By competing with PABA, sulfamethoxazole prevents the initial synthesis of DHF. wikipedia.orgdrugbank.comaap.orgwikipedia.orgsketchy.com
This compound's Action : this compound then targets a subsequent step by competitively inhibiting dihydrofolate reductase (DHFR). This enzyme is essential for reducing DHF to tetrahydrofolic acid (THF), the biologically active form of folate. wikipedia.orgaap.orgsketchy.comwikipedia.orgmims.comnih.govdrugbank.com
This sequential blockade effectively halts the production of THF, which is vital for the synthesis of purines, thymidine, and methionine, all necessary for bacterial DNA and protein production. jove.comwikipedia.orgdrugbank.comsketchy.com While this compound alone is generally bacteriostatic, its combination with sulfamethoxazole can result in bactericidal activity. drugbank.com
The combined use of this compound and sulfamethoxazole has been shown to slow the development of bacterial resistance compared to using either drug alone. drugbank.commsdvetmanual.com This is attributed to the dual-target mechanism, making it more challenging for bacteria to develop resistance to both drugs simultaneously. However, despite this advantage, increased bacterial resistance to this compound-sulfamethoxazole has been observed, particularly with long-term use, such as in prophylaxis for Pneumocystis jirovecii pneumonia in HIV-infected patients. oup.comoup.comresearchgate.netnih.gov Resistance mechanisms can include alterations to the bacterial cell wall, overproduction of DHFR, or the production of resistant DHFR enzymes. drugbank.comtandfonline.com The prevalence of resistance genes, such as sul genes (for sulfonamide resistance) and dfr genes (for this compound resistance), has been documented in various bacterial isolates. nih.gov
Simultaneous Binding to DHFR
Interactions with Other Antimicrobial Agents
Beyond sulfonamides, this compound can exhibit synergistic interactions with other antimicrobial agents, influencing treatment outcomes and interkingdom dynamics in complex infections.
Studies have demonstrated in vitro synergism between this compound and aminoglycosides like amikacin against various Gram-negative bacilli, including Klebsiella pneumoniae, Serratia marcescens, and Escherichia coli. nih.govasm.org This synergy can lead to a greater reduction in bacterial growth and killing compared to either drug used individually. For instance, serum bactericidal titers were found to be significantly higher with the combination of this compound and amikacin than with either drug alone. nih.gov The mechanism for this synergy is not fully elucidated but may involve the ability of beta-lactams to increase the uptake of aminoglycosides, although this specific interaction with this compound is still under investigation. mdpi.com
The following table illustrates the synergistic inhibitory and bactericidal effects of this compound and amikacin against certain Gram-negative bacteria:
| Organism Tested | Fractional Inhibitory Concentration (FIC) Index (Mean ± SD) | Fractional Bactericidal Concentration (FBC) Index (Mean ± SD) | Percentage Synergistically Inhibited (FIC ≤ 0.25) | Percentage Synergistically Killed (FBC ≤ 0.25) |
| Klebsiella pneumoniae | 0.59 ± 0.19 asm.org | 0.55 ± 0.17 asm.org | 40% asm.org | 50% asm.org |
| Serratia marcescens | 0.48 ± 0.18 asm.org | 0.54 ± 0.29 asm.org | 80% asm.org | 65% asm.org |
| Escherichia coli | 0.60 ± 0.22 asm.org | 0.61 ± 0.22 asm.org | 46% asm.org | 46% asm.org |
Note: Synergy was defined as an FIC or FBC index equal to or less than 0.5 in some studies nih.gov, while others used a threshold of ≤ 0.25 for strong synergy asm.org.
This compound can influence interkingdom interactions, particularly in dual infections involving bacteria and fungi. Research has shown that this compound exposure can alter the production of small-molecule natural products by both bacteria and fungi in co-cultivation. For example, in studies involving Burkholderia cenocepacia (bacterium) and Aspergillus fumigatus (fungus), this compound affected the biochemical pathway for DHN-melanin biosynthesis in A. fumigatus, altering fungal conidia pigmentation. researcher.lifeacs.orgacs.org Additionally, this compound was observed to inactivate fragin, a bacterially produced antifungal compound, by A. fumigatus. researcher.lifeacs.org These findings highlight how therapeutic compounds like this compound can shape microbial and fungal metabolomes, influencing interkingdom interactions and the expression of virulence factors in complex infection environments. researcher.lifeacs.orgacs.org
Environmental Research of Trimethoprim
Environmental Fate and Degradation Mechanisms
Trimethoprim (TMP) is generally considered recalcitrant to biodegradation in standard ready and inherent biodegradability tests nih.gov. However, it can undergo degradation through various aerobic and anaerobic biological mechanisms, as well as physico-chemical transformations in surface waters nih.gov.
Biodegradation in Wastewater Treatment Plants (STPs)
The removal of this compound in wastewater treatment plants (STPs) is highly variable, with reported degradation/removal rates ranging from -128% to over 99% across 63 STPs worldwide nih.gov. The average removal efficiency is approximately 25.0%, with a median of 30.0% nih.gov. Minor removal is typically observed during inadequate primary and secondary treatment processes nih.gov. However, the efficiency of TMP biodegradation significantly improves in the presence of nitrifying sludge and with longer sludge retention times (SRTs) in secondary treatment nih.govut.ac.ir. For instance, studies have shown removal efficiencies of 30% for SRTs of 16 and 33 days, increasing to 87% for SRTs of 60-80 days scielo.br. Anaerobic degradation of TMP can also vary from low to rather high efficiencies, depending on conditions nih.goviwaponline.com. Biological nutrient removal (BNR) based sewage treatment processes, such as anaerobic-anoxic-oxic (A2O) and anoxic-oxic (AO) systems, generally exhibit higher antibiotic removal efficiencies compared to oxidation ditch (OD) processes ut.ac.ir.
Table 1: this compound Removal Efficiencies in Wastewater Treatment Plants
| Process Type / Condition | Removal Efficiency (%) | Reference |
| Average (across 63 STPs) | 25.0 (median 30.0) | nih.gov |
| SRT 16-33 days | 30 | scielo.br |
| SRT 60-80 days | 87 | scielo.br |
| Anaerobic conditions | Variable (low to high) | nih.goviwaponline.com |
| A2O / AO processes | Relatively higher | ut.ac.ir |
| Oxidation Ditch (OD) | Relatively lower | ut.ac.ir |
Photodegradation in Aquatic Environments (Direct and Indirect)
This compound degrades slowly under natural solar illumination, with direct photolysis contributing approximately 10% degradation in 500 minutes in demineralized water and up to ~2% in 72 hours in natural water nih.gov. The quantum yield for TMP transformation via direct photolysis is notably low, typically less than 0.002 mdpi.comsemanticscholar.orgscilit.com.
Indirect photolysis plays a more significant role in the degradation of TMP in aquatic environments researchgate.netnih.govrsc.orgsemanticscholar.org. This process is primarily driven by reactions with photosensitizing species, such as hydroxyl radicals (HO•) and triplet excited effluent organic matter researchgate.netnih.govrsc.org. In wastewater effluents, for example, direct photolysis accounts for only about 18% of TMP's photodegradation, while reactions with hydroxyl radicals contribute approximately 62%, and triplet excited effluent organic matter contributes about 20% researchgate.net. The pH of the aquatic environment also influences photodegradation, with higher degradation observed at pH 4.1 compared to pH 7.2 researchgate.net.
Hydrolysis and Sorption Characteristics
This compound is considered stable to hydrolysis, with a reported half-life of one year at 25°C mdpi.com. Its sorption characteristics are generally low to moderate, implying that it predominantly remains in the aqueous phase in environmental waters nih.govnih.gov. The estimated organic carbon-water partition coefficient (Koc) for this compound is 75, suggesting high mobility in soil nih.gov. This compound has a pKa of 7.12, meaning it partially exists in its protonated form in moist soils, and cationic forms tend to adsorb more strongly to soil particles than neutral compounds nih.gov. In sediment cores, the half-lives of this compound were approximately 100 days under anaerobic conditions and 75 days under aerobic conditions, indicating slow biodegradation in these matrices nih.gov.
Role of Environmental Factors (pH, Dissolved Organic Matter, Temperature)
Environmental factors significantly influence the fate and degradation of this compound:
pH: pH plays a crucial role in TMP's degradation and sorption. Photodegradation rates are higher at lower pH values (e.g., pH 4.1 vs. 7.2) researchgate.netmdpi.com. In terms of sorption, TMP is more prone to sorb at higher environmental pH (e.g., pH 9) where it is predominantly non-dissociated, compared to lower pH (e.g., pH 6) where it is mostly dissociated nih.gov. For advanced oxidation processes like Photo-Fenton, optimal degradation of TMP occurs in acidic conditions, typically between pH 3.0 and 4.5 mdpi.com.
Dissolved Organic Matter (DOM): The presence and nature of dissolved organic matter can significantly affect TMP's degradation. While triplet excited effluent organic matter can enhance TMP degradation during indirect photolysis researchgate.net, natural organic matter (NOM) can decrease the degradation rate in UV/persulfate systems nih.gov. Humic acids, a component of DOM, have also been shown to inhibit the ozonation degradation of TMP, possibly due to competitive degradation mdpi.com.
Temperature: Temperature is a primary environmental factor impacting the microbial breakdown of antibiotics, including this compound researchgate.net. Increased temperatures have been correlated with increased degradation rates in aquatic photodegradation, showing an increase of up to ~15% at pH 4 and 7 nih.gov.
Occurrence and Distribution in Environmental Samples
This compound is frequently detected in various environmental samples globally, often in the nanogram per liter (ng/L) to microgram per liter (µg/L) range nih.gov. Its ubiquitous presence is largely attributed to incomplete removal in conventional wastewater treatment processes mdpi.commdpi.com.
Reported concentrations of this compound in different environmental matrices include:
Untreated Municipal Wastewaters: Concentrations typically range from 0.17 µg/L to 8.8 µg/L diva-portal.org.
Hospital Effluents: Can contain higher concentrations, up to 11.3 µg/L diva-portal.org.
Surface Waters: Concentrations can reach up to 0.48 mg/L (480 µg/L) in general surface waters mdpi.com, and up to 28 µg/L in surface waters contaminated by industrial discharges diva-portal.org. In the Thames catchment, elevated concentrations reaching 2000 ng/L (2 µg/L) have been observed, with one specific site recording 3000 ng/L (3 µg/L) biorxiv.org.
Wastewater Treatment Plant Effluents: The highest mean concentration found in Finnish WWTP effluent was 532 ng/L oup.com. In South India, TMP was present at concentrations up to 1 µg/L in STP outlets researchgate.netnih.gov.
Urban Lakes: In Hanoi, Vietnam, the average concentration of this compound in surface water was 8.93 ng/L, with a detection frequency of 100% and a range of 0.65 to 67.9 ng/L semanticscholar.org.
Nationwide Studies (China): A review of antibiotic distribution in China's aquatic environment reported a highest concentration of 924.79 ng/L for TMP in 2009 mdpi.com.
Other Samples: this compound has also been detected in Swedish treated wastewater, surface water, and fish janusinfo.se.
Table 2: Reported Concentrations of this compound in Environmental Samples
| Environmental Compartment | Concentration Range / Mean (µg/L or ng/L) | Reference |
| Untreated Municipal Wastewaters | 0.17 – 8.8 µg/L | diva-portal.org |
| Hospital Effluents | Up to 11.3 µg/L | diva-portal.org |
| Surface Waters (general) | Up to 480 µg/L | mdpi.com |
| Surface Waters (industrial impact) | Up to 28 µg/L | diva-portal.org |
| Thames Catchment Rivers | Up to 3000 ng/L (3 µg/L) | biorxiv.org |
| Finnish WWTP Effluent (mean) | 532 ng/L | oup.com |
| South Indian STP Outlets | Up to 1 µg/L | researchgate.netnih.gov |
| Hanoi Urban Lakes (mean) | 8.93 ng/L (range 0.65-67.9 ng/L) | semanticscholar.org |
| China Nationwide (highest) | 924.79 ng/L | mdpi.com |
Contribution to Antibiotic Resistance in the Environment
This compound, like other antibiotics, is a significant contributor to the selection, maintenance, and increase of antibiotic resistance in environmental bacteria nih.govjanusinfo.seexeter.ac.uk. The presence of even low concentrations of antibiotics in the environment can trigger the development of antibiotic resistance janusinfo.se.
The primary mechanism of this compound resistance in bacteria is the acquisition of mobile dfrA genes frontiersin.orgasm.orgresearchgate.netnih.govsciforum.net. These genes encode variants of the dihydrofolate reductase (DHFR) enzyme that are insensitive to this compound, bypassing the drug's inhibitory effect frontiersin.orgasm.org. dfrA genes are frequently found as gene cassettes within mobile genetic elements such as class 1 and class 2 integrons, as well as transposons and plasmids frontiersin.orgsciforum.net. This association with mobile genetic elements facilitates the rapid horizontal transfer and spread of this compound resistance among diverse bacterial species in the environment frontiersin.orgsciforum.net.
Wastewater treatment plants (WWTPs) are considered crucial hotspots for the dissemination of antibiotic resistance into the environment researchgate.net. They provide favorable conditions for the proliferation of antibiotic-resistant bacteria and for the horizontal transfer of antibiotic resistance genes (ARGs) among different microorganisms researchgate.net. Studies have indicated that concentrations of this compound found in Swedish wastewater treatment plants are likely to select for bacterial resistance janusinfo.se. Consequently, this compound has been added to the Water Framework Directive's Hazardous Compounds Watch List, signifying its potential threat to environmental and human health due to its role in driving resistance exeter.ac.uk. Research has shown a correlative relationship between the residue of TMP and the prevalence of sulfonamide antibiotic resistance genes, such as sul1, in river environments downstream of WWTPs biorxiv.orgresearchgate.net. To mitigate the selection for resistance in aquatic environments, a protective exposure limit of 1 µg/L for this compound has been proposed diva-portal.org.
Impact on Antibiotic-Resistant Genes (ARGs) and Mobile Genetic Elements (MGEs)
This compound contamination in environmental settings has been directly linked to the enrichment and spread of antibiotic-resistant genes (ARGs) and mobile genetic elements (MGEs) researchgate.net. Wastewater treatment plants (WWTPs), in particular, are recognized as critical hotspots for the dissemination of ARGs into the environment biorxiv.org.
Research indicates that the presence of this compound contributes to the increased abundance of specific ARGs, notably the dfrA genes, which directly confer resistance to this compound mdpi.combiorxiv.org. These genes are frequently found in conjunction with sul genes, which confer resistance to sulfonamides, often co-located on the same plasmids or within class 1 integrons (CL1s) mdpi.combiorxiv.org. This co-occurrence highlights the potential for co-selection, where exposure to one antibiotic like this compound can select for resistance to other antibiotics.
The horizontal gene transfer (HGT) of ARGs, facilitated by MGEs such as plasmids and integrons, is a crucial mechanism for the spread of resistance in aquatic environments mdpi.comujms.net. Environmental studies have detected various ARGs relevant to this compound, including dfrE, dfrB2/3, and dfrB1/6, alongside sulfonamide resistance genes like sul1, sul2, sul3, and sul4 biorxiv.org. The sul1 gene, due to its prominence, has frequently been incorporated into the structure of class 1 integrons biorxiv.org.
Despite efforts in wastewater treatment, the reduction in ARG abundance in treated effluents can be lower than anticipated, even when this compound concentrations are significantly reduced biorxiv.org. This suggests that WWTPs may not fully mitigate the risk of ARG dissemination. The abundance of ARGs and MGEs is generally higher in environments impacted by human activities, such as urban and agriculturally polluted areas, compared to pristine environments researchgate.net.
The following table summarizes key ARGs and MGEs associated with this compound resistance in environmental contexts:
| Gene Type | Specific Genes/Elements | Associated Resistance | Environmental Relevance |
| ARGs | dfrA (e.g., dfrA12) | This compound | Directly confers resistance; often plasmid-borne. mdpi.combiorxiv.org |
| ARGs | dfrE, dfrB2/3, dfrB1/6 | This compound | Detected in environmental metagenomes. biorxiv.org |
| ARGs | sul1, sul2, sul3, sul4 | Sulfonamides | Frequently co-occur with dfrA genes, indicating co-selection. mdpi.combiorxiv.org |
| MGEs | Plasmids | Various ARGs | Key vectors for horizontal gene transfer of resistance. mdpi.comujms.net |
| MGEs | Class 1 Integrons (intI1) | Various ARGs | Often carry sul1 and dfrA genes, facilitating their dissemination. mdpi.combiorxiv.org |
Selection Pressure for Resistance Development in Environmental Microorganisms
This compound acts as a significant selective pressure for the development of antibiotic resistance in environmental microorganisms, even at concentrations considerably lower than those used clinically biorxiv.orgbiorxiv.orgmdpi.com. The introduction of antibiotics into the environment, particularly through wastewater discharges, creates conditions that favor the survival and proliferation of resistant bacteria mdpi.comnih.gov.
Studies have demonstrated that exposure to this compound can select for resistant Escherichia coli strains. Notably, resistance development has been observed even when E. coli is exposed to this compound concentrations as low as one-tenth of the established acceptable daily intake (ADI) threshold researchgate.net. This highlights the potential for sub-inhibitory concentrations of antibiotics in the environment to drive resistance evolution.
The evolutionary response of bacteria to this compound can vary depending on the selective pressure. High antibiotic pressures tend to favor the development of strong resistance, while lower pressures might lead to compensatory mutations that mitigate fitness costs associated with resistance biorxiv.org. For instance, early adaptation of E. coli to this compound can involve the de-repression of PhoPQ signaling and the transcriptional upregulation of dihydrofolate reductase (DHFR), the direct target of this compound, often preceding mutations in the folA gene itself biorxiv.org.
Environmental monitoring has revealed elevated concentrations of this compound in various aquatic systems, particularly downstream of wastewater treatment plants, with levels reaching up to 2000 ng/L in some river sites biorxiv.org. Such concentrations are sufficient to exert selective pressure on microbial communities, promoting the persistence and spread of resistance.
The presence of antibiotic residues in aquatic environments is strongly correlated with the occurrence of sul and dfrA genes, especially when these genes are located on mobile genetic elements like plasmids and integrons mdpi.com. This continuous environmental exposure contributes to the maintenance and dissemination of this compound resistance within diverse microbial populations.
The table below illustrates the selective potential of this compound concentrations observed in environmental contexts:
| Environmental Setting | This compound Concentration | Observed Impact on Resistance | Citation |
| Rivers (downstream of WWTPs) | Up to 2000 ng/L | Correlated with prevalence of ARGs. biorxiv.org | |
| Aquatic Biofilms | 100 µg/L | Significantly increased this compound-resistant E. coli cells. researchgate.net | |
| Aquatic Biofilms | 1/10th of ADI dose | Induced selection of this compound-resistant E. coli. researchgate.net |
Novel Applications and Research Directions
Repurposing Trimethoprim for Non-Antimicrobial Applications
Drug repurposing, the identification of new uses for existing drugs, offers a promising strategy for developing potent and less toxic therapeutic agents. This compound is being investigated for applications beyond its conventional antimicrobial role. researchgate.net
Recent in silico studies have explored this compound's potential as a colchicine-binding site (CBS) inhibitor, suggesting its repurposing as an anticancer agent. This compound shares structural similarities and pharmacophoric features with colchicine and combretastatins, which are potent antimitotic agents targeting the CBS. researchgate.netresearchgate.net
Molecular docking studies have shown that this compound exhibits a good binding affinity to the CBS, with an average CDOCKER_ENERGY of -33.75 kcal/mol. researchgate.netresearchgate.net Molecular dynamics simulations, conducted over 100 nanoseconds, further confirmed the stability of the this compound-tubulin complex, with a root mean square deviation (RMSD) of less than 2.5 Å for the protein backbone. researchgate.netresearchgate.net The binding energy, calculated using the Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) approach, was -27.3 kcal/mol, corroborating the docking results. researchgate.netresearchgate.net These findings suggest that this compound has the potential to inhibit tubulin polymerization, a key process in cell division, thereby exhibiting anticancer properties. researchgate.net
Table 1: Computational Binding Data of this compound to Colchicine-Binding Site
| Metric | Value | Reference |
| CDOCKER_ENERGY (average) | -33.75 kcal/mol | researchgate.netresearchgate.net |
| RMSD (protein backbone) | < 2.5 Å (over 100 ns) | researchgate.netresearchgate.net |
| MM-PBSA Binding Energy | -27.3 kcal/mol | researchgate.netresearchgate.net |
Synthesis and Characterization of Metal Complexes and Nanoparticles with this compound
This compound's molecular structure, particularly its pyrimidine ring and amino groups, allows it to act as a chelating agent for metal ions. researchgate.net This property has led to the synthesis and characterization of various metal complexes and nanoparticles incorporating this compound, with investigations into their enhanced biological activities.
Complexes of this compound with transition metals such as Cu(II), Zn(II), Pt(II), Ru(III), Fe(III), Cr(III), Mn(II), Co(II), Cd(II), and Hg(II) have been synthesized. researchgate.netjscimedcentral.comijcsi.projscimedcentral.com Characterization techniques, including UV-Vis, IR, mass spectrometry, and 1H NMR spectroscopy, elemental analysis, electrochemical studies, and thermal analysis, have been employed to elucidate their structures and properties. researchgate.netjscimedcentral.comijcsi.projscimedcentral.comnih.govnsps.org.ngresearchgate.net
These studies have revealed that this compound can act as a monodentate or bidentate ligand, coordinating with metal ions through its amino groups and pyrimidine nitrogen atoms. researchgate.netjscimedcentral.comnih.gov For instance, spectral studies indicate that this compound can link with metal ions as a bidentate ligand through the nitrogen atom N(7) of the amino group and the nitrogen atom N(2) of the pyrimidine ring. jscimedcentral.com Proposed geometries for these complexes include square planar, tetrahedral, and octahedral. researchgate.netjscimedcentral.comijcsi.pronih.govmdpi.com
Furthermore, this compound derivatives and their metal complexes have been functionalized with nanoparticles, such as zinc oxide (ZnO) and gold (Au) nanoparticles. researchgate.netmdpi.com The synthesis of nano-sized complexes of this compound with Fe(II) and Cu(II) using sonication methods has also been reported, with resulting nanocomplexes having sizes of 57.56 nm and 69.88 nm, respectively. nsps.org.ng These nanoparticle functionalizations aim to enhance properties for drug delivery, therapy, and imaging. mdpi.com
Table 2: Examples of this compound Metal Complexes and Nanoparticle Functionalizations
| Metal Ion/Nanoparticle | Coordination Mode (if specified) | Proposed Geometry (if specified) | Reference |
| Cu(II), Zn(II), Pt(II), Ru(III), Fe(III) | Monodentate/Bidentate | Square planar, tetrahedral, octahedral | researchgate.netjscimedcentral.comnih.gov |
| Cr(III), Mn(II), Co(II), Ni(II), Cd(II), Hg(II) | Bidentate | Octahedral | jscimedcentral.comijcsi.projscimedcentral.com |
| ZnO Nanoparticles | Functionalized with complexes | - | researchgate.netmdpi.com |
| Gold (Au) Nanoparticles | Functionalized with complexes | - | researchgate.netmdpi.com |
| Fe(II) nano-sized complex | Through methylene and amino groups | Trigonal bipyramid | nsps.org.ngnsps.org.ng |
| Cu(II) nano-sized complex | Through methylene and amino groups | Trigonal bipyramid | nsps.org.ngnsps.org.ng |
Targeting Human DHFR with this compound Derivatives
While this compound primarily targets bacterial DHFR with high selectivity, research is ongoing to develop this compound derivatives that can effectively inhibit human dihydrofolate reductase (hDHFR). mdpi.commdpi.comresearchgate.netnih.gov Inhibiting hDHFR is a promising strategy for cancer therapy, as this enzyme is essential for cellular processes, including DNA synthesis in rapidly proliferating cells like cancer cells. mdpi.comresearchgate.netnih.gov
Studies have shown that structural modifications to this compound, such as incorporating amide bonds or methylene bridges, can improve binding to the active site of hDHFR and maintain interactions with key residues like Glu-30 and Phe-34. mdpi.com Several synthesized this compound analogs have demonstrated greater hDHFR inhibitory activity than this compound itself. mdpi.commdpi.comnih.gov For example, a series of benzamide this compound derivatives showed hDHFR IC50 values ranging from 4.72 to 20.17 µM, which is significantly more active than this compound's IC50 of 55.26 µM. mdpi.comnih.gov Derivatives like JW2 and JW8 were identified as particularly promising, with JW2 interacting strongly with Gly-117 and JW8 with Asn-64 and Arg-70. mdpi.comnih.gov Other analogs, such as compounds 2 and 3, exhibited hDHFR IC50 values of 0.99 µM and 0.72 µM, respectively, highlighting their enhanced inhibitory properties compared to standard this compound. mdpi.comnih.gov
Table 3: Inhibitory Activity of this compound Derivatives against Human DHFR
| Compound/Derivative | IC50 against hDHFR (µM) | Key Interactions (if specified) | Reference |
| This compound (TMP) | 55.26 | - | mdpi.commdpi.comnih.govnih.gov |
| Benzamide derivatives (JW1-JW8, MB1, MB3, MB4) | 4.72 - 20.17 | - | mdpi.comnih.gov |
| JW2 | (within 4.72-20.17 range) | Gly-117 | mdpi.comnih.gov |
| JW8 | (within 4.72-20.17 range) | Asn-64, Arg-70 | mdpi.comnih.gov |
| Analog 2 | 0.99 | Glu-30 | mdpi.comnih.gov |
| Analog 3 | 0.72 | Glu-30 | mdpi.comnih.gov |
Investigation of this compound's Influence on Host-Pathogen Interactions
Beyond its direct antimicrobial action, this compound has been investigated for its influence on host-pathogen interactions, including potential immunomodulatory effects. tandfonline.comcm-uj.krakow.pl While primarily known for inhibiting bacterial dihydrofolate reductase (DHFR) and thus bacterial nucleic acid synthesis, there are reports suggesting that this compound, particularly in combination with sulfamethoxazole (TMP-SMX), can exert anti-inflammatory and immunomodulatory effects. tandfonline.comcm-uj.krakow.pl
For instance, TMP-SMX has been reported to have a beneficial effect on phyto-hemagglutinin-induced lymphocyte transformation. tandfonline.com Studies have indicated that this compound, at therapeutically relevant dosages, can suppress anti-inflammatory responses. tandfonline.com The combination of this compound and sulfamethoxazole has shown more potent suppressant effects than either drug alone. tandfonline.com
Furthermore, research has explored how this compound exposure can influence chemical interactions between pathogens. In the context of cystic fibrosis, this compound has been shown to induce secondary metabolite production in Burkholderia and affect interkingdom interactions between B. cenocepacia and Aspergillus fumigatus. acs.org This suggests that this compound can modulate the metabolic profiles of pathogens, potentially influencing the resolution of infections. acs.org
In an immunodeficient mouse model of melanoma skin cancer, TMP-SMX significantly improved survival rates, reduced tumor weight and growth, and decreased vascular endothelial growth factor levels. nih.gov This anti-cancer effect was linked to the triggering of an allergic reaction and the promotion of immunity, with TMP-SMX increasing the infiltration of mast cells and the release of allergy-related and immune-enhancing mediators. nih.gov
Q & A
Q. What experimental models are most effective for studying trimethoprim’s antimicrobial efficacy, and how can reproducibility be ensured?
Methodological Answer:
- Use standardized in vitro susceptibility testing (e.g., broth microdilution) with quality-controlled bacterial strains (e.g., E. coli ATCC 25922) to ensure consistency .
- For in vivo models, consider sea urchin embryos to assess developmental toxicity, but restrict concentration ranges (e.g., ≤450 µM) to avoid confounding mortality effects. Document environmental variables (pH, temperature) rigorously .
- Validate reproducibility via inter-laboratory calibration, especially for endpoints like radialization rates or MIC values .
Q. How can researchers optimize LC-MS protocols for quantifying this compound and its metabolites in biological samples?
Methodological Answer:
- Construct calibration curves using ≥20 gradient concentrations of this compound, validated with internal standards. Include quality controls (QCs) to assess intra-day and inter-day precision .
- For metabolite identification, combine MassQL queries (to screen MS2 fragmentation patterns for this compound substructures) with manual validation to reduce false positives. For example, MassQL identified 33/47 candidate features as true this compound derivatives in a recent study .
Q. What statistical approaches are recommended for analyzing dose-response relationships in this compound toxicity studies?
Methodological Answer:
- Use nonlinear regression models (e.g., sigmoidal curves) to fit dose-response data, accounting for inhomogeneous variances. Tools like GraphPad Prism or R’s drc package are suitable .
- Apply principal component analysis (PCA) to FT-IR spectral data to differentiate metabolic responses under varying pH conditions .
Advanced Research Questions
Q. How can network analysis and chemical-genomic datasets elucidate this compound’s synergistic mechanisms with other antibiotics?
Methodological Answer:
- Leverage synthetic lethality concepts: Identify gene pairs whose co-inhibition amplifies this compound’s effect. For example, this compound + sulfamethizole disrupts folate biosynthesis via metabolite accumulation, while this compound + AZT targets distinct pathways .
- Use E. coli K12 chemical-genomic data to predict "two-hit" synergies. Validate hypotheses with growth inhibition assays under combinatorial drug exposure .
Q. What experimental designs mitigate confounding factors when studying this compound resistance reversibility in bacterial populations?
Methodological Answer:
- Conduct longitudinal studies with control cohorts (e.g., a Swedish intervention reduced this compound use by 85% but saw no resistance decline in E. coli). Include phenotypic (e.g., clonal analysis) and genotypic (e.g., dfr gene profiling) metrics .
- Account for co-resistance patterns (e.g., this compound resistance linked to fluoroquinolone resistance) by stratifying isolates using whole-genome sequencing .
Q. How can advanced metabolomics techniques improve the detection of this compound biotransformation products in complex matrices?
Methodological Answer:
- Combine MS2LDA (to extract Mass2Motifs) with MassQL for substructure-specific queries. For instance, MassQL identified a this compound derivative (m/z 549.129) missed by MS2LDA, demonstrating enhanced sensitivity .
- Integrate stable isotope labeling (e.g., ¹³C-trimethoprim) to track metabolic fate in microbial communities or host tissues .
Q. What biochemical assays are critical for characterizing novel this compound-resistant dihydrofolate reductase (DHFR) variants?
Methodological Answer:
- Perform enzyme kinetics (Km, Vmax) using purified DHFR variants under physiological pH (5–7). For example, DfrB7 showed altered substrate affinity compared to wild-type DHFR .
- Use X-ray crystallography or cryo-EM to resolve structural changes in mutant DHFRs that confer this compound resistance .
Contradictory Data Resolution
Q. Why do some studies report pH-dependent this compound efficacy while others show no significant variation?
Methodological Answer:
- Conflicting results may arise from differences in bacterial species (e.g., E. coli vs. Streptococcus pyogenes) or experimental conditions. Standardize pH buffers (e.g., MES for pH 5, HEPES for pH 7) and pre-acclimate cultures to target pH levels before exposure .
- Re-evaluate historical data using multivariate tools like PC-DFA to isolate pH-specific metabolic responses from noise .
Q. How should researchers address discrepancies in this compound resistance gene prevalence across geographical regions?
Methodological Answer:
- Conduct meta-analyses of resistance gene databases (e.g., CARD, ResFinder), stratifying by region and host species. For example, dfrA1 predominates in Europe, while dfrB7 is emerging in Asia .
- Use machine learning (e.g., random forests) to model resistance gene spread, incorporating variables like antibiotic usage patterns and horizontal gene transfer rates .
Methodological Best Practices
Q. What guidelines ensure rigor in reporting this compound-related research?
Methodological Answer:
- Follow the FINER criteria (Feasible, Interesting, Novel, Ethical, Relevant) when formulating research questions .
- Adhere to journal-specific formatting (e.g., ≤3 heading levels, metric units) and statistical reporting standards (e.g., define "significant" only with p-values) .
- Deposit raw data (e.g., LC-MS spectra, resistance gene sequences) in public repositories like MetaboLights or NCBI .
Retrosynthesis Analysis
AI-Powered Synthesis Planning: Our tool employs the Template_relevance Pistachio, Template_relevance Bkms_metabolic, Template_relevance Pistachio_ringbreaker, Template_relevance Reaxys, Template_relevance Reaxys_biocatalysis model, leveraging a vast database of chemical reactions to predict feasible synthetic routes.
One-Step Synthesis Focus: Specifically designed for one-step synthesis, it provides concise and direct routes for your target compounds, streamlining the synthesis process.
Accurate Predictions: Utilizing the extensive PISTACHIO, BKMS_METABOLIC, PISTACHIO_RINGBREAKER, REAXYS, REAXYS_BIOCATALYSIS database, our tool offers high-accuracy predictions, reflecting the latest in chemical research and data.
Strategy Settings
| Precursor scoring | Relevance Heuristic |
|---|---|
| Min. plausibility | 0.01 |
| Model | Template_relevance |
| Template Set | Pistachio/Bkms_metabolic/Pistachio_ringbreaker/Reaxys/Reaxys_biocatalysis |
| Top-N result to add to graph | 6 |
Feasible Synthetic Routes
Featured Recommendations
| Most viewed | ||
|---|---|---|
| Most popular with customers |
Disclaimer and Information on In-Vitro Research Products
Please be aware that all articles and product information presented on BenchChem are intended solely for informational purposes. The products available for purchase on BenchChem are specifically designed for in-vitro studies, which are conducted outside of living organisms. In-vitro studies, derived from the Latin term "in glass," involve experiments performed in controlled laboratory settings using cells or tissues. It is important to note that these products are not categorized as medicines or drugs, and they have not received approval from the FDA for the prevention, treatment, or cure of any medical condition, ailment, or disease. We must emphasize that any form of bodily introduction of these products into humans or animals is strictly prohibited by law. It is essential to adhere to these guidelines to ensure compliance with legal and ethical standards in research and experimentation.


