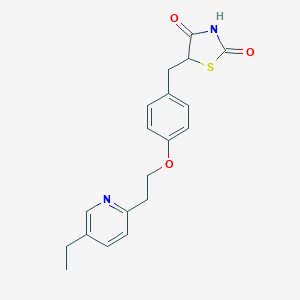
Pioglitazone
Description
Historical Context of Thiazolidinediones and Insulin Sensitization Research
The class of chemical compounds known as thiazolidinediones (TZDs), also referred to as "glitazones," emerged as a novel therapeutic approach for type 2 diabetes in the late 1990s chemrxiv.orgnih.gov. Their introduction represented a distinct departure from existing oral hypoglycemic agents, primarily due to their unique mechanism of action focused on improving insulin sensitivity rather than stimulating insulin secretion conicet.gov.arnih.gov.
The initial research and developmental efforts for thiazolidinediones largely originated in Japan, with ciglitazone (PubChem CID: 2750) being the first compound identified newdrugapprovals.orgconicet.gov.arscienceopen.com. While ciglitazone demonstrated efficacy in improving glycemic control in animal models of insulin resistance, its progression to human trials was hindered by toxicity concerns conicet.gov.arscienceopen.comnih.gov. This setback, however, propelled further research, leading to the development of other, less toxic TZD compounds conicet.gov.arnih.gov.
The activation of PPARγ by TZDs mediates its effects largely by promoting the redistribution of surplus fatty acids to peripheral fat stores. This action reduces the concentration of free fatty acids in the circulation, as well as in the liver and muscle, thereby enhancing insulin sensitivity in these crucial tissues conicet.gov.arnih.gov. Beyond direct fatty acid redistribution, TZDs also modify the secretion of adipokines, which are hormones produced by adipocytes. Notably, they substantially increase the production of adiponectin, a hormone known to enhance fat oxidation and improve insulin action, further contributing to their insulin-sensitizing effects ctdbase.orglabsolu.caconicet.gov.arnih.gov.
Evolution of Pioglitazone within the Thiazolidinedione Class
This compound (PubChem CID: 4829) emerged as a significant member of the thiazolidinedione class, developed by Takeda Pharmaceutical Co., Ltd. newdrugapprovals.orgwikipedia.org. Its entry into the market followed that of troglitazone (PubChem CID: 5591), which was approved in 1997, and alongside rosiglitazone (PubChem CID: 77999), both were approved in 1999 wikipedia.org.
Table 1: Key Thiazolidinedione Compounds and Their Initial Market Approval Years
| Compound | PubChem CID | Initial Market Approval Year (USA) |
| Troglitazone | 5591 | 1997 |
| Rosiglitazone | 77999 | 1999 |
| This compound | 4829 | 1999 |
This compound functions as a potent and highly selective agonist for PPARγ, while also exhibiting a lesser degree of agonistic activity towards PPARα researchgate.netuni.luwikipedia.orgwikipedia.org. This dual action allows this compound to influence both glucose and lipid metabolism comprehensively researchgate.netwikipedia.orgwikipedia.org. Its pharmacological effect primarily involves modulating the transcription of genes that control glucose and lipid production, transport, and utilization in insulin-sensitive tissues such as adipose tissue, skeletal muscle, and the liver researchgate.netuni.lu.
Through this mechanism, this compound effectively reduces insulin resistance in both peripheral tissues and the liver, concurrently decreasing hepatic gluconeogenesis researchgate.netuni.lu. Research findings indicate that this compound increases glucose utilization by skeletal muscle and fat cells, enhances the uptake of free fatty acids, and reduces lipolysis by fat cells mims.comnih.govwikipedia.org. Specific molecular effects include an increase in the expression of glucose transporters 1 and 4 (GLUT-1 and GLUT-4) and an enhancement of insulin signaling pathways mims.comwikipedia.orgwikipedia.org. Moreover, this compound favorably alters the concentrations of various adipokines, notably increasing adiponectin production conicet.gov.arnih.gov. This compound is administered as a racemic mixture, with its enantiomers readily interconverting in vivo and showing no significant pharmacological difference nih.govuni.lu. Its chemical formula is C₁₉H₂₀N₂O₃S nih.govwikipedia.orgresearchgate.net.
Table 2: Key Effects of PPARγ Activation by this compound
| Mechanism of Action | Metabolic Outcome | Relevant Findings |
| PPARγ Agonism | Increases insulin sensitivity in adipose tissue, skeletal muscle, and liver. researchgate.netwikipedia.orguni.lu | Promotes redistribution of fatty acids to peripheral fat. conicet.gov.arnih.gov Reduces circulating fatty acids in liver and muscle. conicet.gov.arnih.gov |
| Gene Transcription Modulation | Alters expression of genes involved in glucose and lipid metabolism. researchgate.netwikipedia.orguni.lu | Increases expression of glucose transporters (e.g., GLUT-1, GLUT-4). wikipedia.orgwikipedia.org Enhances insulin signaling. wikipedia.org Reduces hepatic gluconeogenesis. researchgate.netwikipedia.orguni.lu Increases glucose uptake and utilization in peripheral organs. wikipedia.org |
| Adipokine Secretion Modification | Increases production of adiponectin. conicet.gov.arnih.gov | Adiponectin promotes fat oxidation and improves insulin action. conicet.gov.arnih.gov |
| Lipid Homeostasis | Increases uptake of free fatty acids and reduces lipolysis by fat cells. mims.comnih.govwikipedia.org | Modifies adipocyte differentiation. nih.gov Increases expression of genes involved in glycerol-3-phosphate synthesis (e.g., GPDH) and fatty acid availability (e.g., LPL, ACS) in adipose tissue. mims.com |
Properties
IUPAC Name |
5-[[4-[2-(5-ethylpyridin-2-yl)ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione | |
|---|---|---|
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI |
InChI=1S/C19H20N2O3S/c1-2-13-3-6-15(20-12-13)9-10-24-16-7-4-14(5-8-16)11-17-18(22)21-19(23)25-17/h3-8,12,17H,2,9-11H2,1H3,(H,21,22,23) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI Key |
HYAFETHFCAUJAY-UHFFFAOYSA-N | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Canonical SMILES |
CCC1=CN=C(C=C1)CCOC2=CC=C(C=C2)CC3C(=O)NC(=O)S3 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Formula |
C19H20N2O3S | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
DSSTOX Substance ID |
DTXSID3037129 | |
| Record name | Pioglitazone | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID3037129 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
Molecular Weight |
356.4 g/mol | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Physical Description |
Solid | |
| Record name | Pioglitazone | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0015264 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Solubility |
Practically insoluble, 4.42e-03 g/L | |
| Record name | Pioglitazone | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB01132 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Pioglitazone | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0015264 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Color/Form |
Colorless needles from dimethylformamide and water | |
CAS No. |
111025-46-8, 112529-15-4 | |
| Record name | Pioglitazone | |
| Source | CAS Common Chemistry | |
| URL | https://commonchemistry.cas.org/detail?cas_rn=111025-46-8 | |
| Description | CAS Common Chemistry is an open community resource for accessing chemical information. Nearly 500,000 chemical substances from CAS REGISTRY cover areas of community interest, including common and frequently regulated chemicals, and those relevant to high school and undergraduate chemistry classes. This chemical information, curated by our expert scientists, is provided in alignment with our mission as a division of the American Chemical Society. | |
| Explanation | The data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated. | |
| Record name | Pioglitazone [INN:BAN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0111025468 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Pioglitazone | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB01132 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | pioglitazone hydrochloride | |
| Source | DTP/NCI | |
| URL | https://dtp.cancer.gov/dtpstandard/servlet/dwindex?searchtype=NSC&outputformat=html&searchlist=758876 | |
| Description | The NCI Development Therapeutics Program (DTP) provides services and resources to the academic and private-sector research communities worldwide to facilitate the discovery and development of new cancer therapeutic agents. | |
| Explanation | Unless otherwise indicated, all text within NCI products is free of copyright and may be reused without our permission. Credit the National Cancer Institute as the source. | |
| Record name | Pioglitazone | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID3037129 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
| Record name | 5-({4-[2-(5-ethylpyridin-2-yl)ethoxy]phenyl}methyl)-1,3-thiazolidine-2,4-dione | |
| Source | European Chemicals Agency (ECHA) | |
| URL | https://echa.europa.eu/substance-information/-/substanceinfo/100.114.441 | |
| Description | The European Chemicals Agency (ECHA) is an agency of the European Union which is the driving force among regulatory authorities in implementing the EU's groundbreaking chemicals legislation for the benefit of human health and the environment as well as for innovation and competitiveness. | |
| Explanation | Use of the information, documents and data from the ECHA website is subject to the terms and conditions of this Legal Notice, and subject to other binding limitations provided for under applicable law, the information, documents and data made available on the ECHA website may be reproduced, distributed and/or used, totally or in part, for non-commercial purposes provided that ECHA is acknowledged as the source: "Source: European Chemicals Agency, http://echa.europa.eu/". Such acknowledgement must be included in each copy of the material. ECHA permits and encourages organisations and individuals to create links to the ECHA website under the following cumulative conditions: Links can only be made to webpages that provide a link to the Legal Notice page. | |
| Record name | PIOGLITAZONE | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/X4OV71U42S | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
| Record name | Pioglitazone | |
| Source | Hazardous Substances Data Bank (HSDB) | |
| URL | https://pubchem.ncbi.nlm.nih.gov/source/hsdb/7322 | |
| Description | The Hazardous Substances Data Bank (HSDB) is a toxicology database that focuses on the toxicology of potentially hazardous chemicals. It provides information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. The information in HSDB has been assessed by a Scientific Review Panel. | |
| Record name | Pioglitazone | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0015264 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Melting Point |
193-194C, 183-184 °C, Colorless prisms from ethanol, MP: 193-194 °C. Soluble in dimethyl formamide; slightly soluble in ethanol; very slightly soluble in acetone, acetonitrile. Practically insoluble in water; insoluble in ether. /Pioglitazone hydrochloride/, 183 - 184 °C | |
| Record name | Pioglitazone | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB01132 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Pioglitazone | |
| Source | Hazardous Substances Data Bank (HSDB) | |
| URL | https://pubchem.ncbi.nlm.nih.gov/source/hsdb/7322 | |
| Description | The Hazardous Substances Data Bank (HSDB) is a toxicology database that focuses on the toxicology of potentially hazardous chemicals. It provides information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. The information in HSDB has been assessed by a Scientific Review Panel. | |
| Record name | Pioglitazone | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0015264 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Molecular and Cellular Mechanisms of Action of Pioglitazone
Selective Agonism of Nuclear Receptors
The core molecular action of pioglitazone revolves around its function as a selective agonist for specific nuclear receptors drugbank.comwikipedia.orgmims.comresearchgate.nethellobio.com. These receptors are ligand-activated transcription factors that are instrumental in regulating various metabolic pathways researchgate.net.
Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) Activation
This compound's principal molecular target is the peroxisome proliferator-activated receptor gamma (PPARγ) drugbank.compatsnap.comhellobio.comfrontiersin.orgmdpi.combiorxiv.org. The binding of this compound to PPARγ induces a crucial conformational change within its ligand-binding domain, which is essential for the receptor's activation biorxiv.orgjacc.org. This conformational alteration facilitates the dissociation of corepressor proteins and promotes the subsequent recruitment of coactivator proteins, thereby initiating the transcriptional machinery biorxiv.orgjacc.org. This compound is characterized as a potent and highly selective agonist for PPARγ mims.comhellobio.commdpi.com.
PPARγ exists in two primary isoforms, PPARγ1 and PPARγ2, which originate from alternative splicing and distinct promoter usage oup.comphysiology.org. PPARγ1 is widely distributed and expressed across numerous tissues, including the heart, skeletal muscle, kidney, pancreas, and various epithelial tissues researchgate.netoup.com. In contrast, PPARγ2 demonstrates a more restricted expression pattern, being predominantly found in adipose tissue researchgate.netoup.comphysiology.org. Despite these differences in tissue distribution and the presence of an additional 28 to 30 amino acids at the N-terminal of PPARγ2 compared to PPARγ1, experimental studies have indicated that both murine PPARγ1 and PPARγ2 respond to this compound with comparable half-maximal effective concentrations of activation frontiersin.orgoup.com. Furthermore, this compound exhibits a high affinity for both PPARγ1 and PPARγ2 nih.gov.
The activation of PPARγ by this compound leads to a substantial increase in the transcription of insulin-responsive genes, which are pivotal for regulating glucose and lipid production, transport, and utilization drugbank.compatsnap.comdiabetesonthenet.comresearchgate.netmdpi.com. This profound transcriptional modulation underlies this compound's capacity to enhance insulin sensitivity drugbank.compatsnap.comdiabetesonthenet.com.
Specific genes whose expression is notably influenced by this compound-mediated PPARγ activation include:
Glucose Transporter Type 4 (GLUT4): this compound enhances the expression of GLUT4 on the cell surface, thereby facilitating increased glucose uptake into muscle and adipose tissue cells patsnap.comnih.govphysiology.org.
Genes involved in gluconeogenesis: this compound diminishes hepatic glucose production by downregulating the expression of genes associated with gluconeogenesis in the liver patsnap.comdiabetesonthenet.com.
Genes related to lipid metabolism and storage: This category encompasses genes for fatty acid transporter protein, lipoprotein lipase (LPL), fatty acid synthase (FAS), acetyl-CoA synthetase (ACS), glycerol-3-phosphate dehydrogenase (GPDH), and glucokinase nih.govresearchgate.netdiabetesjournals.org. The activation of PPARγ promotes increased glucose uptake by adipocytes, which is subsequently incorporated into triglycerides for storage diabetesjournals.org.
Research findings from studies examining the effect of this compound on the mRNA levels of genes involved in glycerol-3-phosphate synthesis in subcutaneous fat from individuals with type 2 diabetes illustrate this transcriptional modulation researchgate.net:
| Gene Symbol | Function | This compound-Treated (mRNA level, arbitrary units) | Placebo-Treated (mRNA level, arbitrary units) | Statistical Significance (P-value) |
| PEPCK-C | Gluconeogenesis, Glycerol-3-P synthesis | 1.07 ± 0.10 | 0.59 ± 0.09 | < 0.01 |
| GPDH | Glycerol-3-P synthesis | 1.46 ± 0.07 | 1.13 ± 0.07 | < 0.01 |
The mRNA levels of other key lipogenic genes such as FAS, GPDH, and ACS have also been found to be highly correlated with PPARγ expression and increased upon this compound treatment, suggesting a coordinated transcriptional control system for these genes researchgate.netdiabetesjournals.org.
For PPARγ to effectively execute its transcriptional activation functions, it forms an obligate heterodimer with one of the Retinoid X Receptor (RXR) isoforms, which include RXRα, RXRβ, and RXRγ frontiersin.orgmdpi.comnih.govfrontiersin.orgjci.orgresearchgate.net. This PPARγ/RXR heterodimeric complex subsequently binds to specific DNA sequences known as peroxisome proliferator response elements (PPREs) located within the promoter regions of target genes frontiersin.orgmdpi.comnih.govfrontiersin.orgresearchgate.netfrontiersin.org. This binding event is a critical step in initiating the transcription of genes involved in various metabolic processes, including glucose and lipid metabolism mdpi.com.
Transcriptional Modulation of Insulin-Responsive Genes
Peroxisome Proliferator-Activated Receptor Alpha (PPARα) Modulation
While this compound is predominantly recognized for its agonism of PPARγ, it also functions as a weak agonist of human peroxisome proliferator-activated receptor alpha (PPARα) wikipedia.orgresearchgate.netfrontiersin.orgjacc.orgnih.gov. This modulation of PPARα contributes to additional pharmacological effects, particularly in the regulation of inflammatory responses jacc.orgnih.gov. Research indicates that this compound can repress endothelial TNFα-induced vascular cell adhesion molecule-1 (VCAM-1) mRNA expression and induce hepatic IκBα expression in a PPARα-dependent manner researchgate.netjacc.orgnih.govmedrxiv.org. These anti-inflammatory effects mediated through PPARα are observed most notably in endothelial cells jacc.orgnih.gov. It is important to note that the binding affinity and efficacy of this compound for PPARα are significantly lower when compared to its binding to PPARγ researchgate.net.
Other Potential Molecular Targets (e.g., Amine Oxidase [flavin-containing] B)
Beyond its well-established interactions with the PPAR family, this compound has been identified as a specific and reversible inhibitor of human amine oxidase [flavin-containing] B (MAO-B) drugbank.comdrugbank.commdpi.comacs.orgnih.govresearchgate.net. MAO-B is a flavin-dependent mitochondrial enzyme responsible for the oxidative deamination of certain amine-containing neurotransmitters mdpi.comnih.gov. Structural analysis of the enzyme-inhibitor complex has shown that the R-enantiomer of this compound binds within both the substrate and entrance cavities of the MAO-B active site, forming non-covalent interactions with surrounding amino acid residues drugbank.commdpi.comacs.orgnih.govresearchgate.net. This distinct binding mechanism differentiates this compound from many conventional MAO inhibitors, which often act via covalent inhibition and may not exhibit the same degree of isoenzyme selectivity drugbank.comacs.org. This inhibitory effect on MAO-B suggests potential neuroprotective properties for this compound, a mechanism that has been investigated in the context of neurodegenerative conditions such as Parkinson's disease drugbank.commdpi.comacs.orgnih.gov.
Downstream Molecular Pathways and Cellular Effects
The activation of PPARγ by this compound leads to a cascade of downstream molecular pathways and cellular effects that collectively contribute to improved glycemic control and lipid profiles. drugbank.comresearchgate.net
Enhanced Insulin Signaling and Glucose Disposal
This compound significantly enhances tissue sensitivity to insulin, leading to improved glucose uptake and utilization in peripheral tissues, including skeletal muscle and adipose tissue. drugbank.comresearchgate.netmyendoconsult.comtohoku.ac.jppharmacytimes.com This improvement in insulin sensitivity is a cornerstone of its action, as it allows for better clearance of glucose from the bloodstream without increasing pancreatic insulin secretion. drugbank.comresearchgate.nettohoku.ac.jp Studies have shown that this compound can increase insulin-stimulated glucose disposal by 25–56%. oup.com For instance, a 16-week this compound treatment in type 2 diabetic patients resulted in a 38% increase in insulin-stimulated total glucose disposal during high-dose insulin clamps. diabetesjournals.org This enhancement in glucose disposal contributes to lower plasma glucose and HbA1c levels. drugbank.comnih.gov
Table 1: Effects of this compound on Glucose Parameters in Type 2 Diabetic Patients (Mean Changes After Treatment) diabetesjournals.orgnih.gov
| Parameter | Baseline (Mean ± SD) | Post-Treatment (Mean ± SD) | Mean Change | P-value |
| HbA1c (%) | 7.6 ± 1.2 | 6.4 ± 0.6 | -1.2 ± 1.1 | < 0.001 |
| Fasting Plasma Glucose (mg/dl) | 184 ± 15 | 135 ± 11 | -49 ± 12 | < 0.01 |
| Glucose Infusion Rate (mg/kg/min) | 5.0 (median) | 7.1 (median) | +2.1 | < 0.001 |
Reduction of Hepatic Gluconeogenesis
This compound reduces the hepatic production of glucose, a process known as gluconeogenesis. drugbank.comresearchgate.net This effect is crucial in mitigating fasting hyperglycemia, as the liver is a major site of glucose overproduction in individuals with type 2 diabetes. myendoconsult.comdiabetesjournals.org Research indicates that this compound significantly reduces the fasting rate of gluconeogenesis, and this decrease correlates with a decline in fasting plasma glucose concentrations. diabetesjournals.org This reduction in gluconeogenic flux is considered a strong determinant of this compound's antihyperglycemic effect. diabetesjournals.org
Regulation of Glucose Transporters (GLUT1, GLUT4)
This compound has been shown to increase the expression of glucose transporters, particularly GLUT1 and GLUT4. researchgate.netpharmacytimes.comnih.govthieme-connect.compsu.edu GLUT1 is important for basal glucose uptake, while GLUT4 plays a critical role in insulin-mediated glucose uptake in insulin-sensitive tissues like adipocytes and skeletal muscle. pharmacytimes.comnih.govomicsonline.orgnih.gov In studies involving 3T3-F442A cells, this compound treatment led to significant increases in GLUT1 and GLUT4 mRNA abundance (up to 5-fold) and protein levels (up to 10-fold for GLUT1 and 7-fold for GLUT4). nih.gov This enhancement in transporter expression is largely attributed to the stabilization of their messenger RNA transcripts. nih.gov By increasing the presence of these transporters, this compound facilitates glucose uptake into cells, thus improving glucose utilization and storage. myendoconsult.comnih.gov Interestingly, while this compound increases GLUT1 and GLUT4 expression, studies in human muscle have shown that this compound can reverse the overexpression of GLUT5 (a fructose transporter) observed in diabetic muscle, although the role of this effect in insulin sensitization is not fully clear. diabetesjournals.org
Modulation of Lipid Metabolism
This compound reduces plasma free fatty acid (FFA) levels. researchgate.netoup.comdiabetesjournals.orgthieme-connect.comnih.gov Elevated FFA levels are known to contribute to insulin resistance by stimulating gluconeogenesis and impairing hepatic glucose production. diabetesjournals.org By lowering circulating FFA concentrations, this compound indirectly improves insulin sensitivity and glucose metabolism. nih.govnps.org.au This reduction in FFAs can be attributed to increased fatty acid uptake and storage in adipocytes, thereby redirecting lipid burden away from the liver and muscle. nih.govnps.org.au Studies have reported significant decreases in fasting plasma FFA concentration following this compound treatment, for example, from 647 ± 39 μEq/l to 478 ± 49 μEq/l after 16 weeks. diabetesjournals.org
This compound demonstrates favorable effects on the lipid profile by significantly reducing plasma triglyceride (TG) levels and increasing high-density lipoprotein cholesterol (HDL-C) levels. researchgate.netpharmgkb.orgmyendoconsult.comtohoku.ac.jpsochob.clnih.govdiabetesjournals.orgresearchgate.netscispace.com Clinical trials have consistently shown a reduction in plasma triglycerides by 10–25% and an increase in total HDL-C levels by 10–20%, predominantly due to an increase in the larger HDL2 subfraction. sochob.clresearchgate.net For instance, one study reported a decrease in triglycerides from 219.2 ± 34.4 mg/dl to 189.2 ± 33.7 mg/dl, and an increase in HDL-C from 37.2 ± 2.9 to 41.5 ± 3.1 mg/dl. scispace.com Furthermore, this compound has been observed to reduce levels of small dense LDL particles while increasing levels of larger, less atherogenic LDL fractions. sochob.cldiabetesjournals.orgresearchgate.net This beneficial impact on lipid profiles is a critical aspect of this compound's mechanism, contributing to the management of diabetic dyslipidemia. sochob.clnih.govdiabetesjournals.org
Table 2: Changes in Lipid Parameters with this compound Treatment diabetesjournals.orgscispace.com
| Lipid Parameter | Baseline (Mean ± SD) | Post-Treatment (Mean ± SD) | Mean Change | P-value |
| Triglycerides (mg/dl) | 219.2 ± 34.4 | 189.2 ± 33.7 | -29.9 | < 0.001 |
| HDL Cholesterol (mg/dl) | 37.2 ± 2.9 | 41.5 ± 3.1 | +4.3 | < 0.001 |
| Total Cholesterol (mg/dl) | 201.4 ± 29.8 | 203.2 ± 28.9 | +1.8 | < 0.001 |
| LDL Cholesterol (mg/dl) | 153.7 ± 21.1 | 154.7 ± 20.7 | +0.9 | < 0.001 |
The Chemical Compound this compound: Mechanisms of Action and Biological Effects
This compound, a member of the thiazolidinedione (TZD) class of compounds, is a synthetic ligand for peroxisome proliferator-activated receptor-gamma (PPARγ). Its biological effects are primarily mediated through the activation of this nuclear receptor, which plays a crucial role in regulating glucose and lipid homeostasis. Beyond its well-established metabolic actions, this compound exhibits significant anti-inflammatory and immunomodulatory properties, as well as influences mitochondrial function.
This compound acts as a selective agonist at PPARγ, a nuclear receptor highly expressed in insulin-sensitive tissues such as adipose tissue, skeletal muscle, and liver. Activation of PPARγ by this compound leads to changes in the transcription of insulin-responsive genes, thereby influencing glucose and lipid metabolism, transport, and utilization drugbank.comnih.gov. This mechanism contributes to enhanced tissue sensitivity to insulin and a reduction in hepatic glucose production (gluconeogenesis), leading to improved insulin resistance without increasing pancreatic beta cell insulin secretion drugbank.com.
Adipose Tissue Remodeling and Adipocyte Differentiation
This compound's molecular effects are largely attributed to its PPARγ-mediated regulation of adipocyte differentiation and lipid storage, particularly in adipose tissue where PPARγ is highly abundant nih.gov. The compound promotes the differentiation of preadipocytes into mature adipocytes, leading to an increase in the number of smaller, insulin-sensitive adipocytes diabetesjournals.orgresearchgate.netoup.com. This process, known as adipogenesis or hyperplasia, contributes to the beneficial remodeling of adipose tissue by expanding subcutaneous adipose depots and reducing ectopic and visceral fat accumulation nih.govdiabetesjournals.orgoup.com. The increase in smaller adipocytes with higher lipid storage potential is believed to lower circulating free fatty acid levels, which can improve insulin sensitivity in other tissues like skeletal muscle diabetesjournals.org. Studies have identified four distinct sub-distributions of adipocytes in human abdominal subcutaneous adipose tissue: small (~0.02uL), medium (~0.14uL), large (~0.9uL), and very large (~2.5uL) researchgate.net. This compound treatment has been shown to increase the proportion of small adipocytes while decreasing the area under the curve (AUC) for large adipocytes researchgate.net.
Table 1: Adipocyte Size Distributions and this compound's Effects
| Adipocyte Size Category | Baseline Volume (approx.) | Effect of this compound Treatment |
| Small | ~0.02uL | Increased proportion researchgate.net |
| Medium | ~0.14uL | - |
| Large | ~0.9uL | Decreased AUC researchgate.net |
| Very Large | ~2.5uL | Increased proportion and size researchgate.net |
Influence on Sterol Regulatory Element-Binding Protein-1c (SREBP-1c)
Sterol regulatory element-binding protein-1c (SREBP-1c) is a transcription factor that plays a critical role in regulating lipogenic gene expression and fatty acid biosynthesis researchgate.netnih.gov. This compound has been shown to reduce the expression of genes involved in hepatic de novo lipogenesis, including SREBP-1c, and fatty acid uptake and transport nih.gov. Studies indicate that PPARγ agonists can inhibit SREBP-1c promoter activity induced by Liver X Receptor (LXR) oup.com. This inhibition is mediated through the two LXR response elements (LXREs) within the SREBP-1c promoter region, and PPARs can reduce the binding of LXR/RXR to LXRE oup.com.
Anti-inflammatory and Immunomodulatory Effects
Beyond its metabolic actions, this compound exhibits notable anti-inflammatory and immunomodulatory effects, regulating various immune cells and cytokines tandfonline.commdpi.com. It has been recognized as a potential therapeutic candidate for autoimmune disorders, influencing diseases like systemic lupus erythematosus, psoriasis, inflammatory bowel disease, and multiple sclerosis tandfonline.comnih.gov.
Reduction of Tumor Necrosis Factor Alpha (TNFα)
This compound significantly reduces the production and expression of pro-inflammatory cytokines, including Tumor Necrosis Factor Alpha (TNFα) tandfonline.comahajournals.orgoup.comresearchgate.netnih.gov. TNFα is a cytokine implicated in inflammation and insulin resistance nih.govtandfonline.comdiabetesjournals.org. This compound has been observed to suppress TNFα production in ischemic retinas of wild-type mice, and it can ameliorate TNFα-induced insulin resistance by restoring insulin-stimulated 2-deoxyglucose uptake, tyrosine phosphorylation, and protein levels of insulin receptor (IR) and insulin receptor substrate-1 (IRS-1) ahajournals.orgdiabetesjournals.orgnih.gov. This effect on insulin signaling appears to be independent of PPARγ's adipogenic activity diabetesjournals.orgnih.gov. Furthermore, this compound can directly inhibit TNFα expression and secretion from adipocytes nih.gov.
Squelching of Nuclear Co-factors for Nuclear Factor Kappa B (NF-κB)
This compound, through its action on PPARγ, can inhibit the activity of Nuclear Factor Kappa B (NF-κB), a master regulator of inflammatory responses mdpi.comoup.come-century.usnih.govlongdom.orgmdpi.com. One of the proposed mechanisms involves the "squelching" of nuclear co-factors e-century.usnih.govmdpi.com. PPARγ can scavenge transcriptional co-factors, such as steroid receptor co-activator-1 (SRC-1) and cAMP response element-binding protein (CREB) binding protein (CBP/p300) e-century.us. By binding to PPARγ, these co-factors become unavailable for initiating gene expression, thereby downregulating NF-κB-mediated gene expression e-century.us. Additionally, PPARγ can directly interfere with NF-κB's transcription-activating capacity or indirectly regulate proteins that suppress NF-κB activation, such as by inducing IκBα, the main endogenous inhibitor of NF-κB nih.govlongdom.orgmdpi.com.
Modulation of C-reactive protein (CRP) and Adiponectin Levels
This compound has been shown to modulate the levels of C-reactive protein (CRP) and adiponectin, which are important markers of inflammation and insulin sensitivity, respectively ahajournals.orgoup.comdiabetesjournals.orgnih.govscispace.comfrontiersin.org. This compound treatment leads to a significant decrease in circulating CRP concentrations and a significant increase in plasma adiponectin levels diabetesjournals.orgnih.govscispace.com. This increase in adiponectin contributes to the anti-inflammatory and insulin-sensitizing effects of this compound ahajournals.orgoup.com. Adiponectin, an anti-inflammatory adipokine, is often lower in obese individuals with dysregulated immune responses frontiersin.org.
Table 2: Modulation of Inflammatory Markers by this compound (4-week treatment in Type 2 Diabetic Patients)
| Marker | Baseline Concentration (Mean ± SEM) | This compound Treated Concentration (Mean ± SEM) | P-value | Reference |
| C-reactive protein (CRP) | 3.5 ± 0.6 mg/L | 2.6 ± 0.5 mg/L | P=0.01 | nih.govscispace.com |
| Adiponectin | 3.95 ± 0.57 µg/mL | 7.59 ± 0.95 µg/mL | P=0.002 | nih.govscispace.com |
Mitochondrial Metabolism and Biogenesis
Furthermore, this compound stimulates the expression of genes involved in the fatty acid oxidation pathway, such as carnitine palmitoyltransferase-1 (CPT1), malonyl-CoA decarboxylase, and medium-chain acyl-CoA dehydrogenase diabetesjournals.orgnih.gov. The expression of PPAR-alpha (PPARα), a transcriptional regulator of genes encoding mitochondrial enzymes involved in fatty acid oxidation, is also higher after this compound treatment diabetesjournals.orgnih.gov. In skeletal muscle, this compound therapy can restore insulin sensitivity, partly by coordinating the upregulation of genes involved in mitochondrial oxidative phosphorylation (OXPHOS) and ribosomal protein biosynthesis plos.orgnih.gov. This effect may involve an increase in adiponectin, a direct effect on muscle PPARγ, and/or improved insulin action on mitochondrial biogenesis plos.org. This compound's ability to enhance mitochondrial biogenesis and oxidative phosphorylation may also be mediated via PrPC expression and is associated with improved ATP production researchgate.net.
Table 3: Gene Expression Changes Related to Mitochondrial Function with this compound Treatment
| Gene/Factor | Role in Mitochondrial Metabolism | Effect of this compound Treatment (in adipose tissue) | Reference |
| PGC-1α | Mitochondrial biogenesis | Increased expression diabetesjournals.orgnih.gov | diabetesjournals.orgnih.gov |
| Mitochondrial Transcription Factor A (mtTFA) | Mitochondrial biogenesis | Increased expression diabetesjournals.orgnih.gov | diabetesjournals.orgnih.gov |
| Carnitine Palmitoyltransferase-1 (CPT1) | Fatty acid oxidation pathway | Stimulated expression diabetesjournals.orgnih.gov | diabetesjournals.orgnih.gov |
| Malonyl-CoA Decarboxylase | Fatty acid oxidation pathway | Stimulated expression diabetesjournals.orgnih.gov | diabetesjournals.orgnih.gov |
| Medium-Chain Acyl-CoA Dehydrogenase | Fatty acid oxidation pathway | Stimulated expression diabetesjournals.orgnih.gov | diabetesjournals.orgnih.gov |
| PPARα | Transcriptional regulator of fatty acid oxidation enzymes | Higher expression diabetesjournals.orgnih.gov | diabetesjournals.orgnih.gov |
Mitochondrial Metabolism and Biogenesis
Activation of Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1-alpha (PGC-1α)
This compound, as a PPARγ agonist, has been consistently shown to upregulate the expression of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) diabetesjournals.orgnih.govresearchgate.netmdpi.complos.orgscielo.br. This effect has been observed in various contexts, including human subcutaneous adipose tissue, Down Syndrome fetal fibroblasts, skeletal muscle of women with Polycystic Ovary Syndrome (PCOS), and lung tissues diabetesjournals.orgnih.govplos.orgscielo.br.
The activation and increased expression of PGC-1α by this compound drive several crucial downstream cellular processes:
Mitochondrial Biogenesis and Function: PGC-1α is a master regulator of mitochondrial biogenesis, leading to an increased mitochondrial copy number, enhanced mitochondrial DNA content, and improved mitochondrial respiratory capacity diabetesjournals.orgnih.govmdpi.complos.org. Studies have shown that this compound increases factors involved in mitochondrial biogenesis, including mitochondrial transcription factor A (mtTFA) diabetesjournals.org. It also upregulates the expression of genes required for fatty acid oxidation, such as PPAR-α, medium-chain acyl-CoA dehydrogenase (MCAD), carnitine palmitoyltransferase I (CPT-1), and malonyl-CoA decarboxylase (MLYCD) diabetesjournals.org. Furthermore, this compound can restore the mitochondrial network by influencing fission-fusion machinery, evidenced by the upregulation of optic atrophy 1 (OPA1) and mitofusin 1 (MFN1), and downregulation of dynamin-related protein 1 (DRP1) nih.gov.
Transcriptional Control: The upregulation of PGC-1α by this compound can be mediated, in part, by increased phosphorylation of AMPK nih.govresearchgate.net. PGC-1α also coactivates other key transcription factors, such as Nuclear Respiratory Factor 1 (NRF1) and Estrogen-related receptor (ERR)-alpha, which play vital roles in mediating mitochondrial biogenesis and oxidative metabolism mdpi.com.
Table 1: Impact of this compound on PGC-1α and Related Mitochondrial Markers
| Marker | Observed Effect of this compound | Tissue/Model | Source Index |
| PGC-1α mRNA/Protein | Increased expression | Human subcutaneous adipose tissue, DS fibroblasts, skeletal muscle (PCOS), lung tissue | diabetesjournals.orgnih.govplos.orgscielo.br |
| Mitochondrial Copy Number | Increased | Human subcutaneous adipose tissue | diabetesjournals.org |
| mtTFA mRNA | Increased | Human subcutaneous adipose tissue | diabetesjournals.org |
| Fatty Acid Oxidation Genes | Upregulated (PPAR-α, MCAD, CPT-1, MLYCD) | Human subcutaneous adipose tissue | diabetesjournals.org |
| OPA1, MFN1 | Upregulated (restoration of mitochondrial network) | Trisomic fetal fibroblasts (DS model) | nih.gov |
| DRP1 | Downregulated (restoration of mitochondrial network) | Trisomic fetal fibroblasts (DS model) | nih.gov |
| Citrate Synthase Activity | Increased (marker of mitochondrial aerobic capacity) | Human subcutaneous adipose tissue | diabetesjournals.org |
Expression of Superoxide Dismutases (SOD2)
This compound has been demonstrated to influence the expression of Superoxide Dismutases (SODs), particularly SOD2, a crucial mitochondrial antioxidant enzyme. This compound treatment leads to an upregulation of SOD2 mRNA and protein levels, and in some cases, also SOD1 and SOD3 jpp.krakow.plresearchgate.netnih.govresearcherslinks.com.
The mechanism behind this upregulation involves its interaction with PPARγ and the nuclear factor erythroid 2-related factor 2 (Nrf2) pathway. This compound, through PPARγ-dependent activation and subsequently through PPARγ-dependent activation of Nrf2, can enhance the expression of SOD1, SOD2, and SOD3 jpp.krakow.plresearchgate.netresearcherslinks.com. SOD2 (Manganese Superoxide Dismutase, Mn-SOD), primarily localized in the mitochondrial matrix and inner membrane, plays a critical role in cellular antioxidant defense by converting the superoxide anion radical (•O2-) into hydrogen peroxide (H2O2), thereby preventing oxidative damage nih.govmdpi.com. Research findings indicate that this compound's ability to upregulate these antioxidant defense mechanisms contributes to the attenuation of oxidative DNA damage, as observed in models of high-fat diet-induced hepatic oxidative stress nih.gov. This suggests a protective role of this compound against oxidative stress by enhancing the enzymatic antioxidant defense system.
Table 2: Influence of this compound on Superoxide Dismutase (SOD) Expression
| SOD Isoform | Observed Effect of this compound | Mechanism | Tissue/Model | Source Index |
| SOD1 | Upregulated mRNA/protein | PPARγ-dependent activation, Nrf2 pathway | Renal cortex (pre-hypertensive rats), Mouse preimplantation embryo | jpp.krakow.plresearcherslinks.com |
| SOD2 | Upregulated mRNA/protein | PPARγ-dependent activation, Nrf2 pathway | Renal cortex (pre-hypertensive rats), Liver (high-fat diet mice), Mouse preimplantation embryo | jpp.krakow.plnih.govresearcherslinks.com |
| SOD3 | Upregulated mRNA/protein | PPARγ-dependent activation, Nrf2 pathway | Renal cortex (pre-hypertensive rats), Mouse preimplantation embryo | jpp.krakow.plresearcherslinks.com |
Uncoupling Protein 1 (UCP-1) Expression in Adipose Tissue
This compound significantly impacts the expression of Uncoupling Protein 1 (UCP-1) in various adipose tissue depots. Studies have consistently shown that this compound increases UCP-1 expression in human subcutaneous adipose tissue, white adipose tissue (WAT) in mouse models (e.g., db/db mice), and brown adipose tissue (BAT) diabetesjournals.orgnih.govjci.orgnih.govresearchgate.netphysiology.org.
UCP-1 is a key marker of brown adipose tissue and beige adipocytes, playing a crucial role in non-shivering thermogenesis by uncoupling oxidative phosphorylation from ATP synthesis, leading to heat production diabetesjournals.orgnih.govjci.orgnih.govresearchgate.netphysiology.orgresearchgate.net. The induction of UCP-1 by this compound contributes to an enhanced thermogenic capacity physiology.orgresearchgate.net. This "browning" effect on white adipose tissue, where WAT depots acquire characteristics of BAT, is a notable finding, observed in both in vivo and in vitro studies diabetesjournals.orgjci.orgresearchgate.net. The increase in UCP-1 expression is associated with improved glucose and lipid homeostasis. jci.org
Impact on Islet Beta-Cell Health and Maturity Markers
This compound has a demonstrated impact on the health and maturity markers of pancreatic islet beta-cells, contributing to improved beta-cell function and survival.
Improved Function and Survival: In conditions of lipotoxicity, endoplasmic reticulum (ER) stress, and inflammation, this compound has been shown to improve glucose-stimulated insulin secretion in pancreatic beta-cells e-enm.org. It also represses apoptosis, as evidenced by decreased caspase-3 cleavage e-enm.org.
Reduced Stress and Inflammation: this compound reverses palmitate-induced expression of inflammatory cytokines, such as tumor necrosis factor alpha (TNFα), interleukin 6 (IL-6), and IL-1β e-enm.org. Furthermore, it attenuates ER stress, reducing markers like glucose-regulated protein 78 (GRP78), cleaved-activating transcription factor 6 (ATF6), and C/EBP homologous protein (CHOP) e-enm.org. This protective effect is believed to be mediated, in part, by blocking the NFκB pathway e-enm.orgjcrpe.org.
Maturity Markers: Studies in db/db mice have shown that this compound treatment leads to greater expression of insulin and the transcription factor Nkx6.1 in pancreatic beta-cells, with a corresponding reduction in the abundance of the de-differentiation marker Aldh1a3 nih.govnih.govresearchgate.net. Nkx6.1 is a key marker of mature beta-cells, indicating that this compound helps restore and maintain beta-cell identity nih.gov. Additionally, this compound upregulates glucose transporter 2 (Glut2) expression in beta-cells, which is crucial for glucose sensing e-enm.org.
Preservation of Beta-Cell Mass: In animal models, long-term treatment with this compound has been linked to the preservation of beta-cell mass, primarily through the reduction of oxidative stress e-dmj.org. Clinical studies, such as the Actos Now for Prevention of Diabetes (ACT NOW) study, have shown that this compound can significantly improve insulin sensitivity and beta-cell function, contributing to a decreased rate of conversion from impaired glucose tolerance to type 2 diabetes e-dmj.org. Meta-analyses have further indicated that this compound can elevate the absolute value of homeostasis model assessment of β-cells (HOMA-β) plos.org.
Table 3: Effects of this compound on Islet Beta-Cell Health and Maturity
| Marker/Outcome | Observed Effect of this compound | Tissue/Model | Source Index |
| Glucose-Stimulated Insulin Secretion | Improved | Mouse insulinoma 6 (MIN6) cells | e-enm.org |
| Apoptosis | Repressed (decreased caspase-3/PARP activity) | MIN6 cells | e-enm.org |
| Inflammatory Cytokines (TNFα, IL-6, IL-1β) | Reduced expression | MIN6 cells, Pancreatic islets (KK-Ay mice) | e-enm.org |
| ER Stress Markers (GRP78, ATF6, CHOP) | Reduced expression | MIN6 cells, Pancreatic islets (KK-Ay mice) | e-enm.org |
| Insulin Expression | Increased | Pancreatic β-cells (db/db mice) | nih.govnih.govresearchgate.net |
| Nkx6.1 Expression | Increased | Pancreatic β-cells (db/db mice) | nih.govnih.govresearchgate.net |
| Aldh1a3 (de-differentiation marker) | Reduced abundance | Pancreatic β-cells (db/db mice) | nih.govnih.govresearchgate.net |
| Glucose Transporter 2 (Glut2) | Upregulated expression | Pancreatic β-cells (KK-Ay mice) | e-enm.org |
| Beta-Cell Mass | Preserved (animal models) | db/db mice, Zucker diabetic fatty (ZDF) rats | e-dmj.org |
| HOMA-β (β-cell function) | Elevated (clinical studies) | Patients with Type 2 Diabetes | e-dmj.orgplos.org |
Q & A
Q. What is the primary mechanism of action of pioglitazone, and how can it be experimentally validated?
this compound acts as a peroxisome proliferator-activated receptor gamma (PPAR-γ) agonist, enhancing insulin sensitivity by modulating adipocyte differentiation and lipid metabolism. To validate this, researchers can:
Q. How should a clinical trial investigating this compound’s efficacy in type 2 diabetes mellitus (T2DM) be designed?
A robust trial design should incorporate:
- PICOT framework : Define Population (e.g., T2DM patients with insulin resistance), Intervention (this compound dosage), Comparison (placebo or standard care), Outcome (HbA1c reduction), and Time (6–12 months) .
- Stratified randomization to control for covariates like baseline HbA1c and comorbidities .
- Blinding and placebo-controlled protocols to minimize bias .
Q. What biomarkers are critical for assessing this compound’s metabolic effects?
Key biomarkers include:
- Glycemic control : HbA1c, fasting plasma glucose .
- Lipid metabolism : Triglycerides, HDL cholesterol .
- Inflammation : C-reactive protein (CRP), adiponectin levels .
- Hepatic insulin sensitivity : Measured via hyperinsulinemic-euglycemic clamp .
Advanced Research Questions
Q. How can conflicting data on this compound’s association with cancer risks be resolved?
Contradictory findings (e.g., prostate vs. bladder cancer risks) require:
- Network pharmacology : Use STRING and WebGestalt to map this compound’s target pathways (e.g., PPAR-γ, NF-κB) and correlate with oncogenic pathways .
- Genomic analysis : Query cBioPortal for cancer genomics data (e.g., mutations in PPAR-γ-associated genes) .
- Propensity score matching : Adjust for confounders like age, smoking, and diabetes duration in observational studies .
Q. What methodological approaches are optimal for studying this compound’s non-glycemic effects (e.g., on nonalcoholic steatohepatitis, NASH)?
- Histological validation : Liver biopsies to assess steatosis, ballooning necrosis, and fibrosis .
- Imaging : Magnetic resonance spectroscopy (MRS) for hepatic fat quantification .
- Multi-omics integration : Combine transcriptomic data (PPAR-γ targets) with metabolomic profiling of lipid species .
Q. How can researchers address this compound’s cardiovascular risk paradox (benefits vs. side effects like edema)?
- Subgroup analysis : Stratify patients by baseline cardiovascular risk (e.g., Framingham score) .
- Mechanistic studies : Investigate PPAR-γ’s role in fluid retention via renal sodium reabsorption assays .
- Real-world data : Use EHR-linked cohorts to track long-term outcomes like heart failure hospitalization .
Methodological and Analytical Considerations
Q. What statistical methods are recommended for analyzing this compound’s dose-response relationships?
- Non-linear mixed-effects modeling : For pharmacokinetic/pharmacodynamic (PK/PD) data .
- Cox proportional hazards models : For time-to-event outcomes (e.g., cancer incidence) .
- Meta-regression : To explore heterogeneity in systematic reviews (e.g., variable follow-up durations) .
Q. How should confounding factors be controlled in observational studies on this compound?
- Propensity score matching : Balance covariates like age, BMI, and diabetes severity .
- Sensitivity analyses : Exclude high-risk subgroups (e.g., patients with prior cancer) .
- Instrumental variable methods : Address unmeasured confounders using genetic or geographic proxies .
Q. What experimental models are suitable for studying this compound’s neurovascular effects?
- Animal models : Fructose-fed rats to mimic insulin resistance and assess this compound’s impact on angiotensin II-induced vasoconstriction .
- Ex vivo assays : Isolated vessel preparations to measure endothelial function .
Data Management and Reproducibility
Q. How can researchers ensure reproducibility in this compound-related pharmacokinetic studies?
- HPLC validation : Follow ICH guidelines for quantifying this compound and its metabolites (e.g., column temperature: 30°C, mobile phase: acetonitrile-phosphate buffer) .
- Open-data practices : Share raw chromatograms and calibration curves via repositories like Zenodo .
Q. What are best practices for curating this compound-related research data?
- FAIR principles : Ensure data are Findable, Accessible, Interoperable, and Reusable .
- Structured metadata : Include experimental conditions (e.g., dosage, cohort demographics) and assay protocols .
Retrosynthesis Analysis
AI-Powered Synthesis Planning: Our tool employs the Template_relevance Pistachio, Template_relevance Bkms_metabolic, Template_relevance Pistachio_ringbreaker, Template_relevance Reaxys, Template_relevance Reaxys_biocatalysis model, leveraging a vast database of chemical reactions to predict feasible synthetic routes.
One-Step Synthesis Focus: Specifically designed for one-step synthesis, it provides concise and direct routes for your target compounds, streamlining the synthesis process.
Accurate Predictions: Utilizing the extensive PISTACHIO, BKMS_METABOLIC, PISTACHIO_RINGBREAKER, REAXYS, REAXYS_BIOCATALYSIS database, our tool offers high-accuracy predictions, reflecting the latest in chemical research and data.
Strategy Settings
| Precursor scoring | Relevance Heuristic |
|---|---|
| Min. plausibility | 0.01 |
| Model | Template_relevance |
| Template Set | Pistachio/Bkms_metabolic/Pistachio_ringbreaker/Reaxys/Reaxys_biocatalysis |
| Top-N result to add to graph | 6 |
Feasible Synthetic Routes
Featured Recommendations
| Most viewed | ||
|---|---|---|
| Most popular with customers |
Disclaimer and Information on In-Vitro Research Products
Please be aware that all articles and product information presented on BenchChem are intended solely for informational purposes. The products available for purchase on BenchChem are specifically designed for in-vitro studies, which are conducted outside of living organisms. In-vitro studies, derived from the Latin term "in glass," involve experiments performed in controlled laboratory settings using cells or tissues. It is important to note that these products are not categorized as medicines or drugs, and they have not received approval from the FDA for the prevention, treatment, or cure of any medical condition, ailment, or disease. We must emphasize that any form of bodily introduction of these products into humans or animals is strictly prohibited by law. It is essential to adhere to these guidelines to ensure compliance with legal and ethical standards in research and experimentation.


