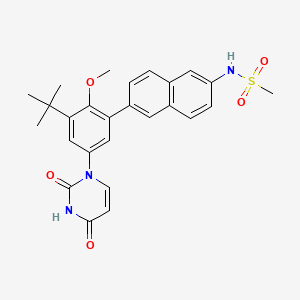
Dasabuvir
Description
Context of Direct-Acting Antiviral Agents (DAAs) in Viral Hepatitis Therapy
Chronic hepatitis C infection is a major global health concern, leading to severe liver diseases such as cirrhosis and hepatocellular carcinoma. plos.org Historically, the treatment of HCV relied on interferon-based therapies, often combined with ribavirin. mdpi.comelsevier.es While these regimens offered some efficacy, they were associated with significant side effects, long treatment durations, and relatively low sustained virological response (SVR) rates, particularly for certain HCV genotypes like genotype 1. mdpi.comnih.gov
The advent of direct-acting antiviral agents marked a paradigm shift in HCV treatment. xiahepublishing.comoup.com DAAs are designed to target specific viral proteins essential for HCV replication, such as the NS3/4A protease, NS5A protein, and NS5B RNA-dependent RNA polymerase. elsevier.esoup.com The introduction of the first-generation DAAs, like the NS3/4A protease inhibitors telaprevir and boceprevir in 2011, significantly improved SVR rates in genotype 1 patients when used in combination with peginterferon and ribavirin. xiahepublishing.comnih.gov However, these regimens still had limitations, including notable adverse effects and the emergence of drug resistance. mdpi.comxiahepublishing.com
The subsequent development of second-generation DAAs, including NS5B polymerase inhibitors like sofosbuvir and non-nucleoside inhibitors such as dasabuvir, further transformed treatment. mdpi.com These newer agents enabled the development of interferon-free regimens, offering higher efficacy, shorter treatment durations, and improved tolerability, ultimately leading to high cure rates (SVR > 90-95%) for many HCV-infected individuals. mdpi.comnih.govracgp.org.aueurekaselect.com
Properties
IUPAC Name |
N-[6-[3-tert-butyl-5-(2,4-dioxopyrimidin-1-yl)-2-methoxyphenyl]naphthalen-2-yl]methanesulfonamide | |
|---|---|---|
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI |
InChI=1S/C26H27N3O5S/c1-26(2,3)22-15-20(29-11-10-23(30)27-25(29)31)14-21(24(22)34-4)18-7-6-17-13-19(28-35(5,32)33)9-8-16(17)12-18/h6-15,28H,1-5H3,(H,27,30,31) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI Key |
NBRBXGKOEOGLOI-UHFFFAOYSA-N | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Canonical SMILES |
CC(C)(C)C1=CC(=CC(=C1OC)C2=CC3=C(C=C2)C=C(C=C3)NS(=O)(=O)C)N4C=CC(=O)NC4=O | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Formula |
C26H27N3O5S | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
DSSTOX Substance ID |
DTXSID301025953 | |
| Record name | Dasabuvir | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID301025953 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
Molecular Weight |
493.6 g/mol | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
CAS No. |
1132935-63-7 | |
| Record name | Dasabuvir | |
| Source | CAS Common Chemistry | |
| URL | https://commonchemistry.cas.org/detail?cas_rn=1132935-63-7 | |
| Description | CAS Common Chemistry is an open community resource for accessing chemical information. Nearly 500,000 chemical substances from CAS REGISTRY cover areas of community interest, including common and frequently regulated chemicals, and those relevant to high school and undergraduate chemistry classes. This chemical information, curated by our expert scientists, is provided in alignment with our mission as a division of the American Chemical Society. | |
| Explanation | The data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated. | |
| Record name | Dasabuvir [USAN:INN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=1132935637 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Dasabuvir | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB09183 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Dasabuvir | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID301025953 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
| Record name | DASABUVIR | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/DE54EQW8T1 | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
Molecular Mechanisms of Action and Target Engagement of Dasabuvir
Elucidation of Hepatitis C Virus NS5B RNA-Dependent RNA Polymerase Inhibition
Dasabuvir is classified as a non-nucleoside inhibitor (NNI) of the HCV NS5B polymerase. drugbank.comnih.govpatsnap.comdrugcentral.orgnih.govmedchemexpress.com Unlike nucleoside inhibitors that mimic natural substrates and are incorporated into the RNA chain, NNIs bind to allosteric sites on the enzyme, leading to conformational changes that impede its function. patsnap.comnih.govresearchgate.net
Non-Nucleoside Binding Site Characterization (Palm I Allosteric Site)
This compound binds to an allosteric site on the HCV NS5B polymerase. patsnap.comnih.govresearchgate.netpatsnap.com Specifically, it targets the palm domain of the enzyme, and is referred to as a non-nucleoside NS5B-palm polymerase inhibitor. drugbank.comdrugcentral.org This binding site is known as the palm I allosteric inhibitory site. nih.gov Resistance mapping studies have indicated that this compound targets this site. drugbank.comdrugcentral.org The palm I site is located within the palm domain of the polymerase and partially overlaps with the palm II site. nih.govasm.org Resistant variants selected by this compound, such as C316Y, M414T, Y448C, Y448H, and S556G, are consistent with binding to the palm I site. nih.gov In silico modeling suggests that this compound binds near the active site, extending from the β-hairpin loop in the thumb domain. nih.gov
Conformational Changes Induced by this compound Binding
Binding of this compound to the NS5B polymerase outside its active site induces a conformational change in the enzyme. drugbank.compatsnap.comnih.govresearchgate.net This allosteric modulation renders the polymerase nonfunctional or unable to elongate viral RNA. drugbank.compatsnap.comnih.govresearchgate.net The induced conformational change prevents the enzyme from effectively carrying out its role in RNA synthesis. drugbank.compatsnap.comnih.govresearchgate.net In silico studies have confirmed that the NS5B-dasabuvir complex is stable and that this compound binding can influence loops involved in the elongation process. nih.gov
Inhibition of Viral RNA Synthesis and Replication
By binding to the allosteric site and inducing conformational changes, this compound effectively inhibits the RNA-dependent RNA polymerase activity of NS5B. drugbank.comwikipedia.orgpatsnap.comdrugcentral.orgcancer.govnih.gov This inhibition prevents the synthesis of new viral RNA molecules, thereby stopping the replication of the HCV genome. drugbank.comwikipedia.orgpatsnap.comcancer.gov The cessation of viral RNA synthesis leads to a reduction in the viral load. patsnap.com
Selective Antiviral Activity Profile
This compound exhibits a selective antiviral activity profile, primarily targeting specific genotypes of the Hepatitis C virus. drugbank.comnih.gov
Genotype-Specific Potency against Hepatitis C Virus (Genotype 1a and 1b)
This compound demonstrates highly potent and selective activity against Hepatitis C virus genotype 1, specifically genotypes 1a and 1b. drugbank.comwikipedia.orgnih.govmedchemexpress.comresearchgate.nettga.gov.auresearchgate.netmedchemexpress.comrxabbvie.comtga.gov.aueuropa.eu Its use is largely limited to genotype 1 infections because the binding sites for non-nucleoside inhibitors like this compound are not well conserved across all HCV genotypes, leading to lower potential for cross-genotypic activity. drugbank.comnih.gov Studies have shown significantly lower potency against polymerases from other HCV genotypes, such as 2a, 2b, 3a, and 4a, often more than 200 times less potent compared to genotype 1. tga.gov.autga.gov.au
Comparative Analysis of Inhibitory Concentrations (IC50 and EC50) in Replicon Systems and Enzymatic Assays
The inhibitory activity of this compound has been quantified through both enzymatic assays using recombinant NS5B polymerase and cell-based replicon assays that measure inhibition of viral RNA replication in cell culture.
In biochemical assays, this compound inhibited recombinant NS5B polymerases derived from HCV genotype 1a and 1b clinical isolates with 50% inhibitory concentration (IC50) values typically ranging between 2.2 and 10.7 nM. nih.govtga.gov.auresearchgate.netmedchemexpress.comrxabbvie.comtga.gov.au The mean IC50 value against a panel of genotype 1a and 1b polymerases was reported as 4.2 nM. tga.gov.autga.gov.aueuropa.eu this compound demonstrated significant selectivity, being at least 7,000-fold more potent against HCV genotype 1 polymerases compared to human/mammalian polymerases. nih.govmedchemexpress.commedchemexpress.com
In HCV subgenomic replicon systems, this compound inhibited genotype 1a (strain H77) and 1b (strain Con1) replicons with 50% effective concentration (EC50) values of 7.7 nM and 1.8 nM, respectively. drugbank.comnih.govmedchemexpress.comtga.gov.aumedchemexpress.comrxabbvie.comtga.gov.aueuropa.euresearchgate.net The activity was retained against a panel of chimeric subgenomic replicons containing NS5B genes from genotype 1 clinical isolates, with EC50s ranging between 0.15 and 8.57 nM. nih.gov The mean EC50 against replicons from treatment-naïve genotype 1a isolates was 0.77 nM (range 0.4 to 2.1 nM), and against genotype 1b isolates was 0.46 nM (range 0.2 to 2 nM). tga.gov.autga.gov.aueuropa.eueuropa.eutga.gov.au
It has been observed that the inhibitory potency of this compound in replicon assays is reduced in the presence of human plasma. A 12- to 13-fold decrease in activity was noted with 40% human plasma, resulting in EC50s of 99 nM for genotype 1a (H77) and 21 nM for genotype 1b (Con1) replicons. nih.govmedchemexpress.comtga.gov.aumedchemexpress.comtga.gov.aueuropa.euresearchgate.net
The M1 metabolite of this compound has also shown antiviral activity, although with lower potency (30-40% less) than the parent compound against genotype 1a and 1b laboratory strains in the replicon assay. tga.gov.autga.gov.au
Table 1: Inhibitory Concentrations of this compound against HCV Genotype 1
Table 2: Selectivity of this compound
| Target | Selectivity Ratio (vs HCV Genotype 1) | Source |
| Human/Mammalian Polymerases | ≥ 7,000-fold selective | nih.govmedchemexpress.commedchemexpress.com |
| HCV Genotypes 2a, 2b, 3a, 4a | > 200 times lower potency | tga.gov.autga.gov.au |
Assessment of Selectivity against Human and Mammalian Polymerases
This compound demonstrates a high degree of selectivity for inhibiting HCV genotype 1 polymerases over a range of human and mammalian polymerases. Studies have shown that this compound is at least 7,000-fold more selective for inhibiting HCV genotype 1 polymerases compared to human/mammalian polymerases. nih.govnih.govmedchemexpress.comselleckchem.comglpbio.com This selectivity was assessed against various human and mammalian DNA-dependent DNA polymerases (including polymerases alpha, beta, and gamma) and DNA-dependent RNA polymerases (such as RNA polymerases II and III), as well as one RNA-dependent DNA polymerase (polymerase gamma-RT function). nih.gov
The inhibitory activity of this compound on recombinant NS5B polymerases derived from HCV genotype 1a and 1b clinical isolates has been measured with 50% inhibitory concentration (IC50) values typically ranging between 2.2 and 10.7 nM. nih.govnih.govmedchemexpress.comglpbio.com In contrast, the activity against human and mammalian polymerases is significantly lower, contributing to its favorable selectivity profile. nih.gov
| Polymerase Type | Selectivity Ratio (this compound: Human/Mammalian Polymerase) |
| HCV Genotype 1 Polymerases | ≥ 7,000:1 |
| Human/Mammalian Polymerases | 1 |
Investigations into Cross-Antiviral Activity beyond Hepatitis C Virus
While primarily developed for HCV, investigations have explored the potential cross-antiviral activity of this compound against other viruses, particularly those within the Flaviviridae family, to which HCV belongs.
Evaluation of this compound Activity against Other Flaviviruses
Studies have evaluated the activity of this compound against other medically important flaviviruses, including Zika virus (ZIKV), West Nile virus (WNV), and tick-borne encephalitis virus (TBEV). mdpi.comresearchgate.netcornell.edunih.gov In in vitro assays using Vero cells, this compound has demonstrated antiviral effects against these flaviviruses at micromolar concentrations. mdpi.comresearchgate.netcornell.edunih.govresearchgate.net
Specifically, this compound exhibited inhibitory effects on the replication of ZIKV (strains MR-766 and Paraiba_01), WNV (strains 13-104 and Eg101), and TBEV (strain Hypr) in Vero cells. mdpi.comresearchgate.netcornell.edunih.gov The 50% effective concentration (EC50) values for this compound against these flaviviruses in Vero cells ranged from 9.09 µM (TBEV) to 10.85 µM (WNV 13-104). mdpi.comresearchgate.netcornell.edu
| Flavivirus Strain | Cell Type | EC50 (µM) |
| ZIKV (MR-766) | Vero | ~10.85 |
| ZIKV (Paraiba_01) | Vero | ~10.85 |
| WNV (13-104) | Vero | 10.85 |
| WNV (Eg101) | Vero | ~15.20-18.82 researchgate.net |
| TBEV (Hypr) | Vero | 9.09 |
This compound also showed anti-ZIKV effects in human astrocytes and a slight effect in neuroblastoma cells, indicating potential cell-type-dependent activity. nih.gov
Analogous Mechanisms of Action in Related Viral Polymerases
This compound functions as a non-nucleoside inhibitor that targets the palm domain of the HCV NS5B polymerase, inducing a conformational change that prevents viral RNA elongation. drugbank.comnih.gov Considering the structural similarities between the RNA-dependent RNA polymerases of HCV and other viruses within the Flaviviridae family, it is hypothesized that the mechanism of action of this compound against these related viral polymerases could be analogous. mdpi.com
While this compound is known to interact with HCV polymerase, the exact binding sites and detailed mechanisms of inhibition for other flaviviruses may vary. mdpi.com The binding sites for non-nucleoside NS5B inhibitors like this compound are reported to be poorly conserved across different HCV genotypes, which contributes to its genotype 1 restriction. drugbank.comnih.gov The extent of conservation of these binding sites in the polymerases of other flaviviruses would influence the analogous nature of this compound's mechanism of action. Despite the structural similarities, the micromolar concentrations required for activity against other flaviviruses, compared to the nanomolar activity against HCV, suggest potential differences in the interaction or binding affinity with the respective polymerases. nih.govmdpi.comresearchgate.netcornell.edunih.gov
Pharmacokinetic and Pharmacodynamic Research of Dasabuvir
Absorption and Distribution Dynamics of Dasabuvir
Research has characterized how this compound is absorbed into the bloodstream and distributed throughout the body.
Absolute Bioavailability Research
Studies have investigated the extent to which this compound is absorbed into the systemic circulation after oral administration. The absolute bioavailability of this compound has been estimated to be approximately 70%. drugbank.comnih.govfda.gov This indicates that a significant portion of the administered dose reaches the bloodstream. Food intake has been shown to increase the exposure (AUC) of this compound. amazonaws.comtga.gov.aueuropa.eu Administration with a moderate-fat meal resulted in a notable increase in this compound AUC and Cmax. europa.eu
Plasma Protein Binding Characteristics
This compound demonstrates high binding to human plasma proteins. It is reported to be greater than 99.5% bound to plasma proteins over a range of concentrations. drugbank.comnih.govamazonaws.comtga.gov.au This extensive protein binding is a significant factor influencing its distribution and elimination. Plasma protein binding of this compound is not substantially altered in patients with renal or hepatic impairment. tga.gov.au
Volume of Distribution Studies
The volume of distribution provides insight into how this compound is distributed between plasma and other body tissues. The volume of distribution of this compound at steady state is reported to be approximately 149 liters. drugbank.comnih.govamazonaws.com Following intravenous administration, the volume of distribution is approximately 400 L. europa.eu The blood to plasma concentration ratio in humans is between 0.5 and 0.7, suggesting preferential distribution in the plasma compartment of whole blood. tga.gov.aueuropa.eu
Here is a table summarizing key absorption and distribution data:
| Parameter | Value | Source(s) |
| Absolute Bioavailability | ~70% | drugbank.comnih.govfda.gov |
| Peak Plasma Concentration (Tmax) | ~4-5 hours | drugbank.comnih.govfda.gov |
| Plasma Protein Binding | >99.5% | drugbank.comnih.govamazonaws.comtga.gov.au |
| Volume of Distribution (Steady State) | 149 L | drugbank.comnih.govamazonaws.com |
| Volume of Distribution (IV) | ~400 L | europa.eu |
| Blood to Plasma Ratio | 0.5 to 0.7 | tga.gov.aueuropa.eu |
Interactive Data Table: Absorption and Distribution of this compound
Metabolic Pathways and Metabolite Characterization of this compound
This compound undergoes significant metabolism, primarily mediated by cytochrome P450 enzymes.
Primary Cytochrome P450 Enzymes Involved (CYP2C8, CYP3A)
This compound is predominantly metabolized by cytochrome P450 (CYP) 2C8. drugbank.comnih.govtga.gov.aufda.govnih.govresearchgate.nethep-druginteractions.orgnih.gov CYP3A also contributes to its metabolism, albeit to a lesser extent. drugbank.comnih.govtga.gov.aunih.govresearchgate.nethep-druginteractions.org This metabolic profile is crucial for understanding potential drug-drug interactions. Strong inhibitors of CYP2C8 can significantly increase this compound exposure, while strong CYP3A inhibitors have a lesser effect. nih.gov Conversely, CYP3A inducers can decrease this compound exposure. nih.gov
Identification and Antiviral Activity of this compound Metabolites (e.g., M1)
Following administration, unchanged this compound is the major component of drug-related radioactivity in plasma, accounting for approximately 60%. tga.gov.au Several metabolites have been identified in plasma, with the most abundant being metabolite M1. tga.gov.auresearchgate.nettga.gov.au M1 represents a significant portion of drug-related radioactivity (around 21% of AUC) in circulation. tga.gov.auresearchgate.nettga.gov.au
Here is a table summarizing key metabolism data:
| Aspect | Details | Source(s) |
| Primary Metabolizing Enzyme | CYP2C8 | drugbank.comnih.govtga.gov.aufda.govnih.govresearchgate.nethep-druginteractions.orgnih.gov |
| Secondary Metabolizing Enzyme | CYP3A (minor contribution) | drugbank.comnih.govtga.gov.aunih.govresearchgate.nethep-druginteractions.org |
| Major Plasma Component | Unchanged this compound (~60% of radioactivity) | tga.gov.auresearchgate.net |
| Most Abundant Metabolite | M1 (~21% of AUC radioactivity) | tga.gov.auresearchgate.nettga.gov.au |
| Primary Biotransformation | Hydroxylation of tert-butyl group to form M1 | researchgate.net |
| M1 Antiviral Activity | Retains antiviral activity, contributes to overall effect | nih.govresearchgate.nettga.gov.au |
| M1 Potency vs this compound | 30-40% lower potency against genotype 1a/1b in replicon assays | tga.gov.autga.gov.au |
| Further M1 Metabolism | Glucuronidation, Sulfation, Secondary Oxidation | researchgate.net |
Interactive Data Table: Metabolism of this compound
Elimination and Excretion Profiles of this compound
The elimination and excretion of this compound have been characterized through various research studies. Understanding these profiles is essential for predicting drug accumulation and potential interactions.
Major Routes of Elimination (Fecal vs. Renal)
Studies indicate that this compound is predominantly eliminated from the body through the feces. Following a single oral dose of [14C]this compound, a mean of 94.4% of the administered radioactivity was recovered in feces. nih.govresearchgate.net In contrast, renal excretion plays a minimal role, with only about 2% of the dose eliminated in the urine. drugbank.comnih.govresearchgate.nettga.gov.au This suggests that hepatic metabolism and subsequent biliary excretion are the primary pathways for this compound clearance. dovepress.com Analysis of excreta revealed that a significant portion of the radioactivity in feces (26.2%) was attributed to the parent compound, while only a negligible amount (0.03%) of the parent compound was found in urine, further supporting metabolism as the major elimination pathway. drugbank.com
Table 1: Major Elimination Routes of this compound
| Route of Elimination | Percentage of Administered Dose |
| Feces | 94.4% |
| Urine | 2.2% (approximately 2%) |
Half-Life Determination and Implications for Dosing Frequency in Research Settings
The elimination half-life of this compound has been determined to be approximately 5.5 to 6 hours. drugbank.comtga.gov.audovepress.commims.com Some research indicates a terminal half-life of approximately 5-8 hours. nih.gov This relatively short half-life supports a twice-daily dosing regimen in clinical research and therapeutic settings to maintain adequate drug concentrations for inhibiting viral replication. nih.gov The M1 metabolite of this compound, which retains antiviral activity, has a half-life similar to that of the parent compound. nih.gov
Table 2: this compound Half-Life
| Parameter | Value |
| Elimination Half-Life | 5.5 to 8 hours |
Pharmacokinetic Modulations by Physiological and Pathophysiological States
The pharmacokinetics of this compound can be influenced by various physiological and pathophysiological conditions, which is important to consider in research study design and interpretation.
Impact of Hepatic Impairment on this compound Exposure
Hepatic impairment has a notable impact on this compound pharmacokinetics, particularly in severe cases. Studies have shown that in subjects with mild or moderate hepatic impairment (Child-Pugh A or B), this compound exposures are minimally affected, with differences in AUC values generally less than 35% compared to individuals with normal hepatic function. tga.gov.autga.gov.aunih.gov However, in subjects with severe hepatic impairment (Child-Pugh C), this compound AUC values were significantly increased, showing a rise of approximately 320% to 325%. tga.gov.aunih.govdovepress.com This substantial increase in exposure in severe hepatic impairment highlights the importance of hepatic function in this compound clearance.
Table 3: Impact of Hepatic Impairment on this compound Exposure (AUC)
| Hepatic Impairment Severity (Child-Pugh Class) | Change in this compound AUC (compared to normal hepatic function) |
| Mild (A) | Minimally affected (e.g., 17% to 24% higher, or 16% to 39% lower) tga.gov.autga.gov.au |
| Moderate (B) | Minimally affected (e.g., 9% to 37% higher, or 16% to 39% lower) tga.gov.autga.gov.au |
| Severe (C) | Significantly increased (approximately 320% to 325%) tga.gov.aunih.govdovepress.com |
Impact of Renal Impairment on this compound Pharmacokinetics
Unlike hepatic impairment, renal impairment does not appear to appreciably alter the pharmacokinetics of this compound. Studies have shown that this compound pharmacokinetic parameters are similar between healthy subjects and those with mild, moderate, or severe renal impairment, including individuals on dialysis. nih.goveuropa.eu Population pharmacokinetic analyses have indicated that creatinine clearance is not a statistically significant predictor of this compound AUC values. researcher.life This is consistent with the minimal renal excretion of this compound. dovepress.com
Influence of Ethnicity on Pharmacokinetic Parameters
Research has investigated the potential influence of ethnicity on this compound pharmacokinetics. Population pharmacokinetic analyses from Phase 3 clinical studies indicated that Asian subjects had approximately 29% to 39% higher this compound exposures (AUC) compared to non-Asian subjects. tga.gov.autga.gov.aueuropa.eu However, other studies suggest that this compound exposures in Asian subjects are comparable with Caucasian subjects. nih.gov While some analyses identified Asian race as a significant covariate for this compound AUC, the magnitude of the effect on drug exposure was considered modest and not clinically significant in some contexts. researcher.lifenih.gov
Pharmacodynamic Relationships and Viral Kinetics
Studies investigating the pharmacodynamic relationships and viral kinetics of this compound, often in combination with other direct-acting antivirals (DAAs), have provided insights into its effect on HCV RNA levels in both plasma and the liver.
Correlation between this compound Exposure and Early Viral RNA Decline
Research has explored the relationship between this compound exposure and the early decline in plasma HCV RNA. In studies involving this compound as part of a multi-DAA regimen, a rapid decline in plasma HCV RNA is observed. For instance, in one study of treatment-naive, non-cirrhotic patients with HCV genotype 1a receiving a regimen including this compound, the mean baseline plasma HCV RNA was 6.7 log10 IU/mL and decreased by -4.1 log10 IU/mL by Day 7 nih.govnih.gov.
However, some analyses have suggested that this compound exposure may not be a significant predictor of sustained virologic response (SVR) in genotype 1a patients when used in combination with other DAAs europa.eu. A simulation study using a semi-mechanistic model of viral dynamics also did not show a significant difference in SVR when the exposures of all components, including this compound, were reduced by 50% europa.eu.
Early viral kinetics, specifically the rate of plasma HCV RNA decline, have been investigated for their potential to predict treatment outcomes or resistance. While some studies have suggested that early viral kinetics can predict DAA resistance, others have not consistently found that viral kinetics during DAA therapy predict SVR nih.govviralhepatitisjournal.org. One study reported that early plasma HCV RNA kinetics predicted treatment responses, but others described detectable plasma HCV RNA levels at the end of therapy in patients who ultimately achieved SVR nih.gov.
Intrahepatic Viral Kinetics in Response to this compound-Containing Regimens
Studies utilizing techniques like fine needle aspiration (FNA) have allowed for the investigation of HCV RNA kinetics within the liver tissue itself, providing a more direct measure of the antiviral effect at the site of infection oup.comoup.comnih.gov.
In a substudy involving HCV genotype 1a-infected individuals coinfected with suppressed HIV receiving a this compound-containing regimen, early events in both liver and plasma were examined. The study found that at baseline, the mean percentage of HCV-infected cells in the liver was 25.2%, which correlated with plasma HCV RNA levels nih.govnih.gov. By Day 7 of treatment, the mean percentage of HCV-infected cells in the liver had significantly decreased to 1.0%, indicating rapid clearance of HCV infection from the liver nih.govnih.gov. DAAs, including this compound, were detectable in the liver tissue at this time point nih.gov.
This rapid decline in infected hepatocytes suggests a strong antiviral effect of the this compound-containing regimen directly within the liver. Based on these findings, researchers extrapolated that HCV eradication in these participants could potentially occur within a relatively short timeframe, estimated to be around 50–63 days of treatment nih.gov. This aligns with the high SVR rates observed in clinical trials of this compound-containing regimens nih.govinfeksiyon.orgplos.orgresearchgate.net.
Comparisons between plasma and intrahepatic viral kinetics have shown that baseline HCV RNA levels were higher and declined more rapidly in plasma than in the liver during the initial phase of treatment oup.comnih.gov. However, the coadministration of ribavirin did not appear to significantly impact the rate of HCV RNA decline in either plasma or liver during the first two weeks of treatment in one study oup.comnih.govnih.gov.
The rapid reduction in infected hepatocytes observed with this compound-containing regimens highlights the potent direct antiviral activity of these therapies at the primary site of HCV replication nih.gov.
Data Table: Early Viral Load Decline
| Study Population (Genotype) | Regimen (includes this compound) | Baseline Mean Plasma HCV RNA (log10 IU/mL) | Mean Change in Plasma HCV RNA by Day 7 (log10 IU/mL) | Mean % HCV-Infected Hepatocytes at Baseline | Mean % HCV-Infected Hepatocytes at Day 7 | Citation |
| Genotype 1a (HIV coinfected) | OBV/PTV/r + DSV ± RBV | 6.7 | -4.1 | 25.2% | 1.0% | nih.govnih.gov |
Note: OBV = Ombitasvir, PTV = Paritaprevir, r = Ritonavir, DSV = this compound, RBV = Ribavirin.
Mechanisms and Landscape of Dasabuvir Resistance
In Vitro Selection and Characterization of Resistance-Associated Substitutions (RASs)
In vitro studies using HCV replicon systems have been crucial in identifying and characterizing the RASs associated with dasabuvir resistance. Passaging replicon-containing cells in the presence of increasing concentrations of this compound selects for variants with reduced susceptibility nih.gov.
Identification of Key Amino Acid Variants in NS5B (e.g., C316Y, S556G, M414T, Y448C/H)
Several key amino acid substitutions in NS5B have been identified through in vitro selection and analysis of treatment-failure isolates that confer resistance to this compound. Prominent among these are substitutions at positions C316, M414, Y448, and S556 nih.gov.
In genotype 1a replicons, S556G was frequently selected at lower this compound concentrations, while C316Y, Y448C, and Y448H were more prevalent at higher concentrations nih.gov. Other variants observed in genotype 1a included M414T nih.gov.
For genotype 1b, common selected variants included C316Y and M414T nih.gov. Substitutions at positions S368, N411, A553, and S556 have also been observed in genotype 1b nih.gov.
Specific RASs identified in NS5B that are associated with this compound resistance include C316Y, S556G, M414T, and Y448C/H nih.gov. Other relevant substitutions include C316N, M414I/V, N411S, Y448C/H, A553T/V, G554S, S556G/R, G558R, D559G/N, and Y561H, depending on the HCV genotype and subtype nih.govtga.gov.au.
Impact of Resistance Mutations on this compound Inhibitory Potency (Fold-Change in EC50/IC50)
The impact of these RASs on this compound's inhibitory potency is typically measured by the fold-change in EC50 (half-maximal effective concentration) or IC50 (half-maximal inhibitory concentration) compared to wild-type virus or enzyme natap.org. A higher fold-change indicates a greater reduction in susceptibility.
Studies have shown varying levels of resistance conferred by different single RASs. For genotype 1a, C316Y and Y448C/H have been reported to confer very high levels of resistance, with fold-changes exceeding 900-fold nih.gov. S556G in genotype 1a resulted in a lower fold-change, typically around 30-fold oup.com. M414T in genotype 1a showed fold-changes around 32-fold oup.com.
In genotype 1b, C316Y and C316N conferred lower levels of resistance compared to genotype 1a C316Y nih.govoup.com. M414T in genotype 1b showed fold-changes around 26-fold oup.com. S556G in genotype 1b conferred approximately 11-fold resistance nih.govoup.com. Some substitutions in genotype 1b, such as S368T and N411S, have shown high fold-changes in resistance nih.gov.
The following table summarizes representative fold-changes in EC50 for selected NS5B RASs in HCV genotype 1:
| HCV Genotype/Subtype | NS5B Variant | Fold-Change in EC50 (vs. Wild-Type) | Source |
| 1a | C316Y | >940, 1472 | nih.govoup.com |
| 1a | M414T | 32 | oup.com |
| 1a | Y448C | >940, 400 | nih.govoup.com |
| 1a | Y448H | >940, 975 | nih.govoup.com |
| 1a | S556G | 30 | oup.com |
| 1b | C316N | 5 | nih.govoup.com |
| 1b | C316Y | 5 | nih.govoup.com |
| 1b | M414T | 26 | oup.com |
| 1b | S556G | 11 | nih.govoup.com |
| 1b | S368T | 1569 | oup.com |
| 1b | N411S | 54 | oup.com |
Combinations of RASs can lead to higher levels of resistance than single substitutions oup.com.
Genetic Barrier to Resistance Across HCV Genotypes
The genetic barrier to resistance refers to the number of nucleotide mutations required for resistance to emerge elsevier.es. For this compound, the genetic barrier is considered to be relatively low, meaning that resistance can potentially arise through a single nucleotide substitution natap.org. This is in contrast to some other classes of antivirals, like nucleoside inhibitors, which often have a higher genetic barrier natap.orgnih.gov.
The low genetic barrier for this compound is attributed to the fact that key resistance-associated amino acid changes can result from a single nucleotide change in the viral RNA sequence egms.de. This facilitates the rapid selection and emergence of resistant variants under drug pressure nih.gov. The emergence of resistance mutations in NS5B can occur via transition or transversion substitutions; transitions generally have a lower energy barrier and occur more frequently nih.gov.
Computational Modeling and Structural Insights into Resistance Mechanisms
Computational modeling techniques, such as molecular docking and molecular dynamics simulations, provide valuable insights into how this compound interacts with NS5B and how RASs affect this interaction to confer resistance nih.govresearchgate.netsemanticscholar.org.
Analysis of this compound-NS5B Binding Pocket Alterations due to Mutations
Computational studies have analyzed the binding pose of this compound within the palm I allosteric site of NS5B and how mutations alter this binding pocket nih.govresearchgate.netsemanticscholar.org. This compound is predicted to bind to the palm I site, extending towards the active site and interacting with residues in the β-hairpin loop (residues L443 to I454) of the thumb domain, as well as residues near the active site in the palm domain nih.govresearchgate.net.
RAS positions, including 316, 414, 448, and 556, are located within or near the this compound binding pocket, and substitutions at these positions can directly impact the shape and properties of the pocket nih.gov. Mutations can cause steric clashes, alter the local environment, or change the flexibility of the binding site, thereby reducing the affinity of this compound for NS5B semanticscholar.org.
For example, computational analysis suggests that mutations can lead to changes in this compound's conformation within the binding pocket semanticscholar.org. Alterations in the binding pocket due to RASs can disrupt key interactions between this compound and NS5B residues that are essential for potent inhibition nih.govsemanticscholar.org.
Changes in Binding Energy and Hydrogen Bonding Interactions
Computational studies can also quantify the impact of RASs on the binding affinity between this compound and NS5B by calculating changes in binding energy semanticscholar.org. A decrease in binding energy (making the value less negative) indicates reduced affinity, consistent with resistance.
Molecular docking and simulation studies have shown that mutations can lead to changes in hydrogen bonding and other non-covalent interactions between this compound and the NS5B protein nih.govsemanticscholar.orgjapsonline.com. This compound has been shown to form hydrogen bonds with residues such as S288, N291, D318, I447, and Y448 in the NS5B binding pocket nih.govresearchgate.net.
Some mutations have been computationally predicted to decrease binding energy, which can be associated with altered hydrogen bonding patterns, including increased or decreased interactions with specific residues or the formation of new hydrogen bonds semanticscholar.org. Conversely, some mutations might increase binding energy, potentially due to decreased interactions with key residues semanticscholar.org. For instance, a computational study indicated that mutations like C316N, C451S, and N411S can decrease binding energy, while M414V, A553V, and C445F might increase it, supported by observed changes in hydrogen bonding and pi-pi interactions semanticscholar.org.
These computational insights help to explain the molecular basis for this compound resistance conferred by specific RASs and can inform the design of new inhibitors aimed at overcoming these resistance mechanisms semanticscholar.org.
Clinical Prevalence and Emergence of this compound Resistance in Patient Cohorts
The emergence of resistance to direct-acting antivirals (DAAs), including this compound, poses a challenge to achieving sustained virologic response (SVR) bmj.com. Resistance-associated substitutions (RASs) can be present at baseline or emerge during or after treatment nih.govoup.com.
Baseline Polymorphisms and Their Clinical Significance
Baseline polymorphisms in the NS5B region associated with this compound resistance have been observed in treatment-naïve patients mdpi.comoup.com. The prevalence of baseline NS5B RASs can vary depending on the HCV genotype and subtype. In genotype 1b (GT1b), baseline NS5B RASs have shown a prevalence of 14.8% in one study mdpi.com. Specifically, the C316N polymorphism is relatively common in GT1b, with reported baseline prevalences ranging from approximately 10% to 36% asm.orgmdpi.com. The S556G variant is also observed in GT1b, with a prevalence of around 11% asm.org.
In genotype 1a (GT1a), baseline NS5B RASs appear to be less common nih.gov. The S556G variant has been reported as the most prevalent baseline NS5B variant in GT1a in some studies asm.org.
Studies have investigated the impact of these baseline polymorphisms on treatment outcome with this compound-containing regimens. In the AVIATOR study, baseline NS5B polymorphisms, including C316N in GT1b and S556G in both GT1a and GT1b, were prevalent, but they did not have a significant impact on the likelihood of achieving SVR when this compound was used in combination with other DAAs asm.orgnih.gov. Similarly, a pooled analysis of Phase 2b and 3 clinical trials indicated that baseline NS5B substitutions/polymorphisms had little impact on SVR rates in GT1a- and GT1b-infected subjects treated with recommended this compound-containing regimens tga.gov.auabbvie.com.
However, the presence of specific baseline NS5B RASs, such as C316N and S556G in GT1b, has been shown to confer reduced susceptibility to this compound in vitro asm.orgoup.com. A combination of C316N and S556G in GT1b can result in a 38-fold reduction in susceptibility to this compound oup.com.
| Genotype/Subtype | NS5B Baseline Polymorphism | Prevalence (%) | In Vitro Fold Resistance (this compound) | Clinical Significance (with combination therapy) |
| GT1b | C316N | ~10-36 | 5 asm.org | Little impact on SVR in clinical trials asm.orgnih.govtga.gov.auabbvie.com |
| GT1b | S556G | ~11 | 11 asm.org | Little impact on SVR in clinical trials asm.orgnih.govtga.gov.auabbvie.com |
| GT1b | C316N + S556G | Not specified | 38 oup.com | Little impact on SVR in clinical trials abbvie.com |
| GT1a | S556G | Not specified | 30 asm.org | Little impact on SVR in clinical trials asm.orgnih.govtga.gov.auabbvie.com |
| GT1a, GT1b | C316Y | Not specified | >900 asm.orgmdpi.com | Associated with resistance in vitro and in treatment failures asm.orgdovepress.comnih.gov |
| GT1a, GT1b | M414T | Not specified | Lower level asm.org | Predominant in GT1b replicon resistance selection; observed in clinical failures asm.orgdovepress.com |
| GT1a | Y448C/H | Not specified | >900 (Y448C/H in GT1b) asm.orgmdpi.com | Associated with resistance in vitro asm.orgmdpi.com |
Development of Resistance During and After this compound-Containing Therapies
The development of resistance during or after treatment with this compound-containing regimens is a key factor in treatment failure dovepress.com. In clinical trials evaluating this compound in combination with other DAAs (such as paritaprevir and ombitasvir, with or without ribavirin), virologic failure rates were low, but resistance-associated substitutions were frequently observed in patients who failed therapy nih.govasm.orgnih.govfda.govelsevier.es.
In GT1a patients who experienced virologic failure, treatment-emergent NS5B RASs included S556G asm.orgnih.govmdpi.comdovepress.com. Other variants selected in vitro or identified in clinical trials conferring resistance to this compound include substitutions at amino acid positions C316, S368, A395, N411, M414, E446 (GT1a only), Y448, A553, G554, S556, or D559 tga.gov.autga.gov.au. The C316Y, M414T, Y448H, and S556G variants have been noted to have higher replication efficiency rates, potentially contributing to resistance dovepress.comnih.gov.
In GT1b patients who failed treatment, treatment-emergent NS5B RASs included S556G mdpi.com. The predominant variants selected in genotype 1b replicon resistance selection were C316Y and M414T asm.org.
The persistence of these treatment-emergent RASs after treatment failure has been assessed. In GT1a-infected subjects in Phase 2b trials, this compound treatment-emergent variants like M414T, G554S, S556G, G558R, or D559G/N in NS5B were observed tga.gov.au. Due to the high SVR rates in GT1b, trends in the persistence of treatment-emergent variants in this genotype could not be established tga.gov.au. The long-term clinical impact of the emergence or persistence of virus containing this compound-resistance-associated substitutions is not fully known tga.gov.au.
Cross-Resistance Profiles with Other Antiviral Agents
Cross-resistance is expected among antiviral agents within the same class that target the same viral protein and binding site fda.govrxlist.com. As a non-nucleoside NS5B-palm polymerase inhibitor, cross-resistance is expected between this compound and other non-nucleoside NS5B-palm polymerase inhibitors fda.govrxlist.com. However, at the time of some reports, no other drugs of this specific class were approved fda.gov.
Cross-resistance is generally not expected between this compound and other classes of DAAs, including NS3/4A protease inhibitors, NS5A inhibitors, and nucleoside/nucleotide NS5B inhibitors, due to their distinct mechanisms of action and binding sites fda.govrxlist.comgutnliver.org. For example, this compound retained full activity against HCV replicons containing single substitutions associated with resistance to other polymerase inhibitors, such as the S282T variant in the nucleoside binding site (associated with sofosbuvir resistance) and variants in the thumb domain (like M423T, P495A, P495S, and V499A) asm.orgrxlist.com.
While cross-resistance is not expected with other DAA classes, treatment failure with a this compound-containing regimen (typically used in combination with other DAAs) can lead to the selection of RASs in multiple viral targets (NS3, NS5A, and NS5B), depending on the specific combination therapy used nih.govfda.govelsevier.es. Over 90% of GT1a subjects experiencing virologic failure with a 12 or 24-week this compound-containing regimen developed RASs in at least one antiviral target, and over 50% developed RASs in all three antiviral targets fda.gov.
| Antiviral Class | Cross-Resistance with this compound | Rationale |
| Non-nucleoside NS5B-palm inhibitors | Expected | Same target (NS5B) and binding site (palm I) fda.govrxlist.com |
| Nucleoside/Nucleotide NS5B inhibitors | Not expected | Different binding site (catalytic site) fda.govrxlist.comgutnliver.org |
| NS3/4A Protease Inhibitors | Not expected | Different viral target (NS3/4A) fda.govrxlist.com |
| NS5A Inhibitors | Not expected | Different viral target (NS5A) fda.govrxlist.com |
| Interferon and Ribavirin | Not expected | Different mechanisms of action fda.gov |
Clinical Research Methodologies and Efficacy Studies of Dasabuvir
Design of Pivotal Clinical Trials and Observational Studies Involving Dasabuvir
Pivotal clinical trials and observational studies have been instrumental in evaluating the efficacy and safety of this compound-based regimens. These studies have employed various designs to assess outcomes in diverse patient groups.
Phase 3 and Phase 3b Study Designs
Phase 3 and 3b studies involving this compound have typically been multicenter trials, often open-label, evaluating the efficacy and safety of this compound in combination with other DAAs, with or without ribavirin (RBV). These studies have enrolled both treatment-naive and treatment-experienced patients with chronic HCV genotype 1 infection, including those with compensated cirrhosis. elsevier.esdovepress.comahdbonline.comresearchgate.net
For instance, the TOPAZ-III study (M14-225) was a Phase 3b, open-label, multicenter trial conducted in Brazil to evaluate a this compound-containing regimen in genotype 1-infected patients with advanced hepatic fibrosis (METAVIR F3-F4). elsevier.es This study included both treatment-naive and interferon-experienced patients. elsevier.esresearchgate.net Similarly, the SAPPHIRE-I and SAPPHIRE-II trials were Phase III studies that assessed this compound-based therapy in treatment-naive and treatment-experienced non-cirrhotic patients with genotype 1 HCV, respectively. nih.govdovepress.com PEARL-II and PEARL-III studies evaluated this compound in combination with other DAAs in genotype 1b patients without cirrhosis. dovepress.comahdbonline.com TURQUOISE-II was an open-label randomized clinical trial that enrolled treatment-naive or treatment-experienced patients with genotype 1a or 1b and compensated cirrhosis. ahdbonline.com GEODE-II was a Phase 3, open-label, multicenter study that evaluated a this compound-containing regimen with low-dose ribavirin in genotype 1a-infected patients without cirrhosis. nih.gov
Study designs have varied in duration, with treatment periods typically ranging from 12 to 24 weeks, and follow-up periods extending to 12 or 24 weeks post-treatment to assess sustained virologic response (SVR). elsevier.esdovepress.comnih.govtga.gov.au Some studies, like TOPAZ-II, have included longer-term follow-up periods (e.g., five years post-treatment) to evaluate the long-term impact of SVR on liver disease progression. abbvie.comabbvie.com While some studies were open-label, the assessment of HCV RNA as an objective laboratory measurement was considered unlikely to introduce bias in efficacy reporting. elsevier.es
Real-World Effectiveness Studies and Patient-Reported Outcomes
Beyond controlled clinical trials, real-world effectiveness studies have provided valuable insights into the performance of this compound-based regimens in broader patient populations encountered in routine clinical practice. elsevier.esinfeksiyon.org These observational studies often include patients with a wider range of comorbidities and characteristics than those typically enrolled in pivotal trials. plos.org
For example, the AMBER study assessed the real-world effectiveness and safety of a this compound-containing regimen in patients with chronic hepatitis C, including individuals with advanced liver disease and those who had not responded to previous therapies. infeksiyon.org Another real-world study in Southern Brazil evaluated the SVR rates among compensated chronic hepatitis C patients treated with various DAA therapies, including a this compound-containing regimen. elsevier.es These studies have generally shown high effectiveness rates consistent with those observed in Phase 3 trials. infeksiyon.orgplos.org
While the provided search results mention patient-reported outcomes in the context of real-world studies, detailed findings on specific patient-reported outcomes related to this compound were not extensively available in the snippets.
Virologic Outcomes Assessment in this compound-Based Therapies
A primary endpoint in studies evaluating this compound-based regimens is the assessment of virologic outcomes, particularly sustained virologic response (SVR).
Sustained Virologic Response (SVR) Rates across Patient Subgroups
Sustained virologic response, typically defined as undetectable HCV RNA at 12 weeks (SVR12) or 24 weeks (SVR24) after the completion of treatment, is the key measure of treatment success. abbvie.cominfeksiyon.org this compound-based regimens have demonstrated high SVR rates across various patient subgroups with HCV genotype 1 infection. nih.govnih.gov
The SVR12 and SVR24 rates are highly concordant, with SVR12 being a strong predictor of SVR24. tga.gov.auabbvie.com
Here is a summary of SVR12 rates in selected subgroups based on the search results:
| Patient Subgroup | Regimen | SVR12 Rate (%) | Source |
| Genotype 1 overall (Phase 3 pooled) | This compound + other DAAs ± RBV | >95 | nih.gov |
| Genotype 1 overall (Treatment-naive, non-cirrhotic) | This compound + other DAAs + RBV (12 weeks) | 96.2 | dovepress.comnatap.org |
| Genotype 1a (Treatment-naive, non-cirrhotic) | This compound + other DAAs + RBV (12 weeks) | 95.3 | nih.govdovepress.com |
| Genotype 1b (Treatment-naive, non-cirrhotic) | This compound + other DAAs + RBV (12 weeks) | 98.0 - 99.0 | nih.govdovepress.com |
| Genotype 1 (Treatment-experienced, non-cirrhotic) | This compound + other DAAs + RBV (12 weeks) | 96.3 | dovepress.com |
| Genotype 1 (Compensated cirrhosis, 12 weeks) | This compound + other DAAs + RBV | 91.8 | dovepress.com |
| Genotype 1 (Compensated cirrhosis, 24 weeks) | This compound + other DAAs + RBV | 95.9 | dovepress.com |
| Genotype 1b (Compensated cirrhosis, 12 weeks) | This compound + other DAAs | 100 | termedia.pl |
| Genotype 1 (Real-world, overall) | This compound + other DAAs ± RBV | 99.0 | infeksiyon.org |
| Genotype 1 (Real-world, cirrhotic) | This compound + other DAAs ± RBV | 98.3 | infeksiyon.org |
Analysis of Virologic Failure and Post-Treatment Relapse
Despite high SVR rates, a small percentage of patients treated with this compound-based regimens may experience virologic failure, either during treatment (breakthrough) or after completing therapy (relapse). abbvie.comnatap.org In Phase 3 trials, the rate of on-treatment virologic failure was low, around 0.2-0.8%. dovepress.comabbvie.comnatap.orgabbvie.com Post-treatment relapse rates were also low, typically ranging from 1.5% to 2.7%. dovepress.comabbvie.comnatap.orgabbvie.com
Analysis of virologic failure has focused on identifying the presence of resistance-associated substitutions (RASs) in the HCV genome. elsevier.esdovepress.comtga.gov.auabbvie.comfda.gov Treatment-emergent RASs have been detected in patients who experienced virologic failure with this compound-based regimens. elsevier.esdovepress.comtga.gov.auabbvie.comfda.gov These substitutions can occur in the NS3, NS5A, and NS5B regions, corresponding to the targets of the combined DAA therapy. elsevier.esdovepress.comtga.gov.auabbvie.comfda.gov
In patients who experienced virologic failure with this compound-containing regimens, NS5B substitutions, including those at positions C316Y, M414T/V, Y448H/C, G554S, S556G, and D559G, have been associated with reduced susceptibility to this compound in vitro. nih.govmdpi.com Analysis of patients who failed this compound-based therapy has shown the emergence of NS5B substitutions, such as M414I and C316N. abbvie.com The presence of RASs at the time of virologic failure is common. elsevier.esdovepress.comtga.gov.auabbvie.comfda.govmdpi.comoup.com
Virologic failure was more prevalent in genotype 1a patients compared to 1b in some studies, potentially linked to resistance patterns. nih.gov Factors associated with treatment failure can include prior treatment experience, presence of cirrhosis, and pre-existing RASs. oup.com
Efficacy Research in Specific Patient Populations
This compound-based regimens have also been evaluated in specific patient populations who historically had limited treatment options or faced additional complexities.
Studies have assessed the efficacy of this compound in combination with other DAAs in liver transplant recipients with recurrent HCV genotype 1 infection. nih.govoatext.comclinicbarcelona.organnalsoftransplantation.comscielo.br These studies have demonstrated high SVR rates in this population, with one real-world study reporting a 100% SVR12 rate in liver transplant recipients with genotype 1b. oatext.comannalsoftransplantation.com The CORAL-I study, a Phase III trial, also found high SVR rates (97%) in liver transplant recipients with genotype 1. oatext.com
Patients coinfected with HCV genotype 1 and HIV-1 have also been studied. nih.govnih.gov The TURQUOISE I trial assessed a this compound-containing regimen plus RBV in this population, achieving high SVR rates. nih.gov
Efficacy has also been investigated in patients with renal impairment. While the provided snippets did not offer detailed efficacy data for this compound specifically in patients with renal impairment, one abstract mentioned a Phase 3b study (MALACHITE-I and MALACHITE-II) that included patients with renal impairment. abbvie.com Another source indicates that no dose adjustment of this compound is required for patients with mild, moderate, or severe renal impairment, suggesting its potential use in this population. europa.eu A real-life cohort study evaluated a this compound-containing regimen in patients with severe renal impairment and end-stage renal disease, reporting high SVR rates. mjima.org
The efficacy of this compound in patients with decompensated cirrhosis (Child-Pugh B or C) is generally not recommended, with its use typically limited to patients with compensated cirrhosis (Child-Pugh A). nih.goveuropa.eu
Treatment-Naive versus Treatment-Experienced Patients
Clinical trials have investigated the efficacy of this compound-containing regimens in both patients who have not received prior HCV treatment (treatment-naive) and those who have failed previous therapies, particularly interferon-based regimens (treatment-experienced). Studies such as MALACHITE-I and MALACHITE-II compared the all-oral combination of ombitasvir/paritaprevir/ritonavir and this compound ± ribavirin (OBV/PTV/r+DSV±RBV) with telaprevir plus pegylated interferon/ribavirin (TPV+PegIFN/RBV) in non-cirrhotic, chronic HCV GT1-infected patients. nih.gov
In MALACHITE-I, which included treatment-naive patients, SVR12 rates with OBV/PTV/r+DSV+RBV were 97% for genotype 1a and 99% for genotype 1b. nih.gov For genotype 1b, the regimen without ribavirin also showed a high SVR12 rate of 98%. nih.gov In contrast, the TPV+PegIFN/RBV regimen resulted in lower SVR12 rates (82% for genotype 1a and 78% for genotype 1b). nih.gov
MALACHITE-II focused on treatment-experienced patients (failure to prior PegIFN/RBV therapy). The OBV/PTV/r+DSV+RBV regimen achieved an SVR12 rate of 99%, significantly higher than the 66% observed with TPV+PegIFN/RBV. nih.gov
These studies demonstrate that this compound, as part of the 3D regimen, provides high SVR rates in both treatment-naive and treatment-experienced patients with chronic HCV genotype 1 infection.
| Patient Population | Regimen | Genotype | SVR12 Rate (%) | Study/Source |
| Treatment-Naive | OBV/PTV/r+DSV+RBV | 1a | 97 | MALACHITE-I |
| Treatment-Naive | OBV/PTV/r+DSV+RBV | 1b | 99 | MALACHITE-I |
| Treatment-Naive | OBV/PTV/r+DSV | 1b | 98 | MALACHITE-I |
| Treatment-Experienced | OBV/PTV/r+DSV+RBV | 1 | 99 | MALACHITE-II |
| Treatment-Naive/Experienced | OBV/PTV/r±DSV±RBV (Pooled) | 1/4 | 96 | Real-world researchgate.net |
Patients with Compensated Cirrhosis and Advanced Fibrosis
The efficacy of this compound-containing regimens has also been evaluated in patients with more advanced liver disease, specifically those with compensated cirrhosis (Child-Pugh A) and advanced fibrosis (METAVIR F3/4).
The TURQUOISE-II trial enrolled treatment-naive or treatment-experienced patients with HCV genotype 1a or 1b and compensated cirrhosis (Child-Pugh A). ahdbonline.com Treatment with ombitasvir, paritaprevir, and ritonavir plus this compound, with ribavirin, for 12 or 24 weeks resulted in high SVR rates. ahdbonline.com
Another real-world study in Romania including patients with F2, F3, and F4 fibrosis (compensated cirrhosis) infected with HCV genotype 1b reported an efficacy rate of over 99% with OBV/PTV/r combined with this compound, with or without ribavirin. researchgate.net
Research has also investigated the regimen in patients with non-compensated cirrhosis (Child-Pugh B or C), though it is generally recommended to be avoided in this population due to potential risks of hepatic decompensation. ejgm.co.uk However, one study in patients with HCV GT1b and non-compensated cirrhosis showed a 100% SVR rate in those treated with the 3D therapy, including those who discontinued early for safety reasons. ejgm.co.uk
| Patient Population | Regimen | Genotype | SVR12 Rate (%) | Study/Source |
| Compensated Cirrhosis (Child-Pugh A) | OBV/PTV/r+DSV+RBV | 1a/1b | High SVR rates | TURQUOISE-II ahdbonline.com |
| Advanced Fibrosis/Comp. Cirrhosis | OBV/PTV/r+DSV±RBV | 1 | 99.2 (Overall) | Real-world Taiwan nih.gov |
| Advanced Fibrosis/Comp. Cirrhosis | OBV/PTV/r+DSV±RBV | 1b | >99 | Real-world Romania researchgate.net |
| Non-Compensated Cirrhosis | OBV/PTV/r+DSV±RBV | 1b | 100 | Real-world Russia ejgm.co.uk |
Patients with Severe Renal Impairment and End-Stage Renal Disease (ESRD)
The efficacy and safety of this compound-containing regimens have been evaluated in patients with chronic kidney disease (CKD), including those with severe renal impairment (eGFR <30 mL/min/1.73 m²) and ESRD, including those on hemodialysis.
An open-label study (RUBY-II) evaluated the efficacy and safety of OBV/PTV/r with or without this compound in adults with HCV genotype 1a or 4 infection and severe renal impairment or ESRD. clinicaltrialsregister.eu
A real-life cohort study in the Czech Republic evaluated the PrOD regimen in 23 patients with HCV GT1 (21 GT1b, 2 GT1a), including 4 with CKD4 and 19 on maintenance hemodialysis. karger.com Six patients had compensated liver cirrhosis, and three were liver transplant recipients. karger.com All 23 patients completed the 12-week treatment and achieved an SVR12 rate of 100%. karger.com
These findings indicate that this compound, as part of the 3D regimen, is highly effective in achieving SVR in patients with severe renal impairment and ESRD infected with HCV genotype 1, particularly genotype 1b.
| Patient Population | Regimen | Genotype | SVR12 Rate (%) | Study/Source |
| Severe Renal Impairment/ESRD | OBV/PTV/r±DSV | 1a/4 | Evaluated | RUBY-II clinicaltrialsregister.eu |
| Severe Renal Impairment/ESRD (Real-life) | PrOD | 1 | 100 | Czech Republic karger.com |
| ESRD (Pooled Analysis) | OBV/PTV/r ± DSV | 1b | 99.8 | Meta-analysis frontiersin.org |
| ESRD (Pooled Analysis) | OBV/PTV/r ± DSV + RBV | 1b | 100 | Meta-analysis frontiersin.org |
| ESRD (Pooled Analysis, mITT) | OBV/PTV/r ± DSV | 1 | 98.31 | Meta-analysis frontiersin.org |
Research in Patients Coinfected with HIV-1
Clinical research has also focused on the efficacy of this compound-containing regimens in patients coinfected with both HCV genotype 1 and HIV-1.
The TURQUOISE-I trial evaluated OBV/PTV/r plus DSV treatment in patients with HIV-1/HCV genotype 1 coinfection, with or without cirrhosis. oup.com Part 1a of this phase 2 trial reported SVR12 rates of 94% and 91% after 12 or 24 weeks of treatment, respectively. oup.com Excluding nonvirologic failures, the SVR12 rate was 98% for genotype 1. oup.com
A real-life study of PrOD ± ribavirin in HIV/HCV-coinfected patients with GT1 in routine clinical practice in Spain reported that 172 out of 182 patients (94%) attained SVR. tandfonline.com The SVR rates by mITT analysis were 97% for genotype 1a and 98% for genotype 1b. tandfonline.comtandfonline.com In this study, 98% of patients with cirrhosis achieved SVR. tandfonline.comtandfonline.com
A comprehensive analysis of the effectiveness of OBV/PTV/r, this compound for HCV in HIV/HCV coinfected subjects, including clinical trials and observational studies, found a pooled estimated SVR12 rate of 96.3% for genotype 1. nih.gov Subgroup analysis showed SVR12 rates of 96.8% for treatment-naive and 98.9% for treatment-experienced patients with genotype 1. nih.gov Patients with cirrhosis had a pooled SVR12 rate of 97.8%, while those without cirrhosis had a rate of 96.7%. nih.gov
These studies demonstrate high SVR rates with this compound-containing regimens in HIV-1 coinfected patients with HCV genotype 1.
| Patient Population | Regimen | Genotype | SVR12 Rate (%) | Study/Source |
| HIV-1 Coinfected | OBV/PTV/r+DSV | 1 | 94-91 | TURQUOISE-I Part 1a oup.com |
| HIV-1 Coinfected (Real-life) | PrOD ± RBV | 1 | 94 (Overall) | Real-life Spain tandfonline.com |
| HIV-1 Coinfected (mITT) | PrOD ± RBV | 1a | 97 | Real-life Spain tandfonline.comtandfonline.com |
| HIV-1 Coinfected (mITT) | PrOD ± RBV | 1b | 98 | Real-life Spain tandfonline.comtandfonline.com |
| HIV-1 Coinfected (Pooled) | OBV/PTV/r ± DSV ± RBV | 1 | 96.3 | Comprehensive Analysis nih.gov |
| HIV-1 Coinfected TN (Pooled) | OBV/PTV/r ± DSV ± RBV | 1 | 96.8 | Comprehensive Analysis nih.gov |
| HIV-1 Coinfected TE (Pooled) | OBV/PTV/r ± DSV ± RBV | 1 | 98.9 | Comprehensive Analysis nih.gov |
| HIV-1 Coinfected Cirrhosis | OBV/PTV/r ± DSV ± RBV | 1 | 97.8 | Comprehensive Analysis nih.gov |
Post-Liver Transplant Patients
Recurrent HCV infection after liver transplantation significantly impacts graft and patient survival. Clinical research has investigated the use of this compound-containing regimens in this specific population.
The CORAL-I study, a multicenter phase III study, evaluated the safety and efficacy of ombitasvir/paritaprevir/ritonavir and this compound with ribavirin for 24 weeks in HCV genotype 1-infected liver transplant recipients who were at least 12 months post-transplant. europa.euoatext.com This study found the therapy to be both safe and effective, with SVR rates of 97% in genotype 1. oatext.com
A real-world study (AMBER-CEE) assessed the efficacy and safety of OBV/PTV/r/+DSV±RBV in the treatment of post-transplant recurrence of HCV genotype 1 infection. researchgate.netannalsoftransplantation.com This study included 35 liver transplant recipients with recurrent HCV genotype 1 infection, the majority of whom were genotype 1b-infected and treatment-experienced. researchgate.netannalsoftransplantation.com SVR12 was achieved by all patients (35/35, 100%), irrespective of prior treatment history or degree of fibrosis. researchgate.netannalsoftransplantation.com
A single-center real-world study in Spain reported a 100% SVR rate at 12 weeks post-treatment with oral ombitasvir combined with ritonavir-paritaprevir plus this compound and ribavirin for 24 weeks in 22 liver graft recipients transplanted due to cirrhosis caused by genotype 1 HCV with post-transplantation viral recurrence and scant fibrosis. oatext.com
These studies indicate that this compound, as part of the 3D regimen, is highly effective in achieving SVR in post-liver transplant patients with recurrent HCV genotype 1 infection.
| Patient Population | Regimen | Genotype | SVR Rate (%) | Study/Source | Timepoint |
| Post-Liver Transplant | OBV/PTV/r+DSV+RBV | 1 | 97 | CORAL-I oatext.com | SVR12 |
| Post-Liver Transplant (Real-world) | OBV/PTV/r/+DSV±RBV | 1 | 100 | AMBER-CEE researchgate.netannalsoftransplantation.com | SVR12 |
| Post-Liver Transplant (Real-world) | OBV/PTV/r+DSV+RBV | 1 | 100 | Single-center Spain oatext.com | SVR12 |
Future Directions and Emerging Research Avenues for Dasabuvir
Development of Novel Dasabuvir Derivatives with Improved Affinities
Research efforts are directed towards designing and developing novel this compound derivatives with enhanced binding affinities to the HCV NS5B polymerase. Computational studies play a significant role in this area, analyzing the effect of mutations on this compound binding and guiding the design of new compounds. For instance, computational mutation analysis on HCV NS5B has shown that certain mutations can alter this compound's conformation and binding energy. Some mutations, like C316N, C451S, and N411S, have been observed to decrease binding energy, while others, such as M414V, A553V, and C445F, can increase it. semanticscholar.org Future work aims to design new this compound derivatives with improved affinity for HCV NS5B, potentially overcoming the impact of resistance-associated mutations. semanticscholar.org Chemoinformatic approaches are also being employed to design derivatives with potentially improved safety and pharmacokinetic profiles. acs.orgnih.govresearchgate.netresearchgate.net
Strategies to Overcome Existing Resistance Profiles
Resistance-associated substitutions (RASs) in the NS5B polymerase can reduce this compound's effectiveness. mdpi.com Key this compound RASs in HCV genotype 1a include C316Y and S556G, while in genotype 1b, they include C316Y and M414T. nih.gov In vitro studies have shown that variants like C316Y in both genotype 1a and 1b, and Y448C/H in genotype 1b, can confer high levels of resistance to this compound. mdpi.com Strategies to overcome existing resistance profiles primarily involve the use of this compound in combination with other DAAs that have non-overlapping resistance profiles. mdpi.comeuropa.eu The combination of this compound with ombitasvir (an NS5A inhibitor) and paritaprevir (an NS3/4A inhibitor), often boosted by ritonavir, has demonstrated a higher barrier to resistance compared to monotherapy. nih.govmdpi.com Research continues to explore optimal combination regimens and the role of baseline RASs in treatment outcomes, particularly in genotype 1a patients. practicalgastro.com
Advanced Predictive Modeling for this compound Efficacy and Resistance
Advanced predictive modeling techniques, including in silico methods, are being utilized to better understand this compound's interaction with NS5B and predict efficacy and the emergence of resistance. Molecular docking and dynamics simulations are used to model this compound's binding pose with the NS5B polymerase and elucidate the reasons behind resistance associated with clinically relevant RASs. nih.govnih.gov These models have shown that this compound binds at the palm-I site of NS5B and interacts with specific residues, and that RAS positions are often located within or near the this compound binding pocket. nih.govnih.gov Such computational studies help in evaluating the influence of structural descriptors on inhibitory bioactivity and guiding the design of more effective molecules with improved binding affinity. d-nb.info Predictive modeling can aid in identifying potential resistance before treatment and informing personalized treatment strategies. researchgate.netresearchgate.net
Role of this compound in Broad-Spectrum Antiviral Development
While primarily known for its activity against HCV, research is exploring the potential of this compound in the development of broad-spectrum antivirals. Given that this compound targets an RNA-dependent RNA polymerase (RdRp), an enzyme essential for the replication of many RNA viruses, its activity against other viruses with similar RdRp structures is being investigated. mdpi.com Studies have shown that this compound can inhibit the infection of other RNA viruses, such as human norovirus (HuNoV), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and human rotavirus A (RVA) in human intestinal enteroids. nih.govresearchgate.netasm.org This suggests that this compound or its derivatives could potentially be explored for activity against a wider range of viral pathogens, highlighting its potential role in broad-spectrum antiviral development. mdpi.comscientificarchives.com
Exploration of this compound in Other Viral Pathologies
Beyond its established use for HCV genotype 1 infection, the potential of this compound is being explored in the context of other viral pathologies. Pre-clinical studies, often employing in silico and in vitro methods, are investigating its activity against other viruses. For example, this compound has been studied for its potential inhibitory effects on the replication of vector-borne flaviviruses, including Zika virus (ZIKV) and West Nile virus, due to similarities in their RdRp enzymes with that of HCV. acs.orgnih.govresearchgate.netmdpi.com These studies suggest that this compound or its derivatives might have inhibitory activity against the RdRP and/or methyltransferase of these viruses. acs.orgnih.gov The demonstration of this compound's activity against HuNoV, SARS-CoV-2, and RVA in human intestinal enteroids further supports the exploration of its potential in treating other viral infections, particularly those affecting the gastrointestinal or respiratory tracts. nih.govresearchgate.netasm.org
Q & A
Q. What is the primary mechanism of action of dasabuvir against hepatitis C virus (HCV)?
this compound acts as a non-nucleoside inhibitor (NNI) of the HCV NS5B RNA-dependent RNA polymerase (RdRp). It binds to the palm domain of NS5B, inducing conformational changes that inhibit viral RNA synthesis. Key evidence includes its EC₅₀ values of 7.7 nM (genotype 1a) and 1.8 nM (genotype 1b) in replicon assays . Methodologically, RdRp inhibition is validated via in vitro polymerase activity assays and subgenomic replicon systems .
Q. How is this compound’s in vitro antiviral activity assessed against HCV genotypes?
Antiviral efficacy is quantified using subgenomic replicon systems transfected into hepatoma cell lines (e.g., Huh-7). Key metrics include:
- EC₅₀ : Determined via dose-response curves (Table 1).
- Selectivity index (SI) : Ratio of cytotoxic concentration (CC₅₀) to EC₅₀.
| Genotype | EC₅₀ (nM) | Reference |
|---|---|---|
| 1a (H77) | 7.7 | |
| 1b (Con1) | 1.8 |
Replicon resistance profiling involves maintaining cells under drug pressure to select for mutations (e.g., C316Y, M414T) .
Q. Which metabolic enzymes are involved in this compound’s clearance?
this compound is metabolized primarily by CYP2C8 (60%) and CYP3A4 (30%). The major metabolite, M1 (hydroxylated tert-butyl), undergoes glucuronidation and sulfation. Methodological validation includes:
Q. What experimental methods quantify this compound and its metabolites in biological matrices?
Stability-indicating HPLC-DAD and UPLC-MS/MS are used:
- HPLC-DAD : Validated for this compound and degradation products (e.g., alkaline DP1/DP2) with LOD 0.12 µg/mL and LOQ 0.37 µg/mL .
- UPLC-MS/MS : Quantifies this compound, M1, and co-administered DAAs (e.g., ombitasvir) in plasma, with deuterated internal standards (e.g., C13D3-dasabuvir) .
Advanced Research Questions
Q. How does this compound’s activity extend to non-HCV viruses (e.g., EV-A71, dengue)?
Repurposing studies use structure-based virtual screening and functional assays:
- EV-A71 : this compound inhibits viral replication (IC₅₀ = 1.8 µM) via ROCK1 interaction, validated by qPCR, TCID₅₀, and cytokine profiling (MCP-1, TNF-α reduction) .
- Dengue : Plaque reduction assays show 1 µM this compound reduces viral titers in co-infection models, though efficacy requires further validation .
Q. What methodologies identify resistance mutations in HCV NS5B after this compound exposure?
Resistance profiling involves:
- In vitro selection : Replicons cultured under escalating this compound concentrations (10–100× EC₅₀) yield mutations (e.g., C316N, S556G) .
- Computational mutagenesis : Molecular dynamics simulations analyze mutation effects on binding affinity (e.g., C445F disrupts hydrogen bonding with Asn291) .
Q. How do structural biology techniques elucidate this compound-NS5B interactions?
- Docking studies : this compound’s binding to the palm I site is modeled using NS5B crystal structures (PDB: 4WTD). Key interactions include hydrophobic contacts with Phe193 and hydrogen bonds with Asn291 .
- Site-directed mutagenesis : Mutant NS5B (e.g., M414V) is expressed in replicons to assess reduced this compound susceptibility (EC₅₀ shifts >10-fold) .
Q. What pharmacokinetic (PK) considerations apply to this compound in hepatic impairment models?
PK studies in cirrhotic patients reveal:
- AUC changes : +325% in CP-C patients (Child-Pugh C) due to reduced CYP2C8 activity .
- Protein binding : Unbound fraction increases from 0.61% (healthy) to 0.42% (CP-C), impacting free drug exposure .
Methodologically, population PK models integrate covariates (e.g., albumin, bilirubin) to predict dose adjustments .
Q. Why is this compound combined with ombitasvir/paritaprevir/ritonavir in HCV therapy?
The regimen targets multiple viral proteins:
Q. How does this compound’s anti-inflammatory activity modulate viral pathogenesis?
In EV-A71-infected THP-1 cells, this compound reduces pro-inflammatory cytokines (MCP-1, TNF-α) via:
Q. Contradictions and Limitations
Q. Methodological Gaps
- In vivo models : Limited data on this compound’s efficacy in animal models of dengue or EV-A71.
- Cross-resistance : Impact of NS5B mutations on other palm-domain inhibitors (e.g., tegobuvir) remains underexplored .
Featured Recommendations
| Most viewed | ||
|---|---|---|
| Most popular with customers |
Disclaimer and Information on In-Vitro Research Products
Please be aware that all articles and product information presented on BenchChem are intended solely for informational purposes. The products available for purchase on BenchChem are specifically designed for in-vitro studies, which are conducted outside of living organisms. In-vitro studies, derived from the Latin term "in glass," involve experiments performed in controlled laboratory settings using cells or tissues. It is important to note that these products are not categorized as medicines or drugs, and they have not received approval from the FDA for the prevention, treatment, or cure of any medical condition, ailment, or disease. We must emphasize that any form of bodily introduction of these products into humans or animals is strictly prohibited by law. It is essential to adhere to these guidelines to ensure compliance with legal and ethical standards in research and experimentation.


