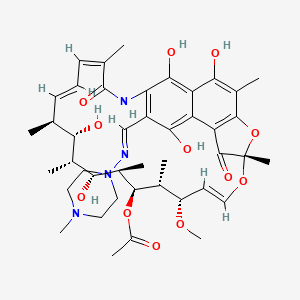
Rifampicin
Description
Properties
Key on ui mechanism of action |
Rifampin acts via the inhibition of DNA-dependent RNA polymerase, leading to a suppression of RNA synthesis and cell death. Although rifampin is most active during cell multiplication ... /it/ appears to have some effect on resting cells. Electron microscopy has revealed changes in cytoplasm and disappearance of ribosomes in tubercle bacilli exposed to rifampin, indicating inhibition of DNA-dependent RNA polymerase. Rifampin inhibits DNA-dependent RNA polymerase of mycobacteria and other microorganisms by forming a stable drug-enzyme complex, leading to suppression of initiation of chain formation (but not chain elongation) in RNA synthesis. More specifically, the beta subunit of this complex enzyme is the site of action of the drug, although rifampin binds only to the holoenzyme. Nuclear RNA polymerase from a variety of eukaryotic cells does not bind rifampin, and RNA synthesis is correspondingly unaffected. While rifampin can inhibit RNA synthesis in mammalian mitochondria, considerably higher concentrations of the drug are required than for the inhibition of the bacterial enzyme. High concentrations of rifamycin antibiotics also inhibit viral DNA-dependent RNA polymerases and reverse transcriptases. Rifampin is bactericidal for both intracellular and extracellular microorganisms. Developmental expression of CYPlAl, CYPlA2 and CYP3A6 in the rabbit have been studied. Cytochromes P450IAl, P450IA2 and P450IIIA6 exhibited comparable patterns of developmental expression. Present at low level (less than 0.05 mnol/ng) in the new born animal up to week 3, these proteins sharply accumulated between weeks 3 and 4 to reach a maximum by week 4 (P450IAl, 0.2 nmol/mg; P450IA2, 0.8 nmol/ng; P450IIIA6, 0.12 nmol/mg) and decr in the adult (P450IAl, 0.2 nmol/mg; P450IA2, 0.4 mnol/mg; P450IIIA6, 0.09 nmol/mg). Cytochromes P450IAl and P450IA2 were not expressed in the untreated fetus. Onset of CYP3A6 gene expression occurred at day 30 of gestation and both transcription and mRNA accumulation were transplacentally inducible by rifampicin only shortly before birth, i.e. after treatment of the females between days 28 and 30 of gestation. Both long (1.85 kb) and short (1.7 kb) mRNA transcripts were expressed in untreated or rifampicin treated fetuses. CYP3A6 gene expression was also induced by rifampicin in l week old and 2 week old animals. Developmental expression of CYPlAl and CYPlA2 genes was shown to be closely related to the diet change accompanying weaning which occurs at weeks 3-4. In animals subjected to either delayed (week 6) or early (week 2) weaning, sharp accumulation of messages, proteins and related activities were delayed or anticipated accordingly with respect to normal weaning. Artificially scheduled weaning gave similar results when repeated with biological grade lucern (grown in the absence of chemical fertilizers, pesticides) ... the main constituent of commercial rabbit chow. While CYP3A6 gene expression could be brought forward by early weaning at week 2, both message and protein did not exhibit incr accumulation after delayed weaning at week 6, and remained at the low level of the new born animal. Treatment of l week old and 2 week old animals with triiodothyronine or of 3 week old animals with propylthiouracil, an antithyroid factor, did not modify the normal pattern of developmental expression of genes CYPlAl, CYPlA2 and CYP3A6. ... |
|---|---|
CAS No. |
13292-46-1 |
Molecular Formula |
C43H58N4O12 |
Molecular Weight |
822.9 g/mol |
IUPAC Name |
[(7S,11S,12R,13S,14R,15R,16R,17S,18S,19Z,21Z)-2,15,17,27,29-pentahydroxy-11-methoxy-3,7,12,14,16,18,22-heptamethyl-26-[(E)-(4-methylpiperazin-1-yl)iminomethyl]-6,23-dioxo-8,30-dioxa-24-azatetracyclo[23.3.1.14,7.05,28]triaconta-1(29),2,4,9,19,21,25,27-octaen-13-yl] acetate |
InChI |
InChI=1S/C43H58N4O12/c1-21-12-11-13-22(2)42(55)45-33-28(20-44-47-17-15-46(9)16-18-47)37(52)30-31(38(33)53)36(51)26(6)40-32(30)41(54)43(8,59-40)57-19-14-29(56-10)23(3)39(58-27(7)48)25(5)35(50)24(4)34(21)49/h11-14,19-21,23-25,29,34-35,39,49-53H,15-18H2,1-10H3,(H,45,55)/b12-11-,19-14?,22-13-,44-20+/t21-,23+,24+,25+,29-,34-,35+,39+,43-/m0/s1 |
InChI Key |
JQXXHWHPUNPDRT-CHTVYFSYSA-N |
SMILES |
CC1C=CC=C(C(=O)NC2=C(C(=C3C(=C2O)C(=C(C4=C3C(=O)C(O4)(OC=CC(C(C(C(C(C(C1O)C)O)C)OC(=O)C)C)OC)C)C)O)O)C=NN5CCN(CC5)C)C |
Isomeric SMILES |
C[C@H]1/C=C\C=C(/C(=O)NC2=C(C(=C3C(=C2O)C(=C(C4=C3C(=O)[C@](O4)(OC=C[C@@H]([C@H]([C@H]([C@@H]([C@@H]([C@@H]([C@H]1O)C)O)C)OC(=O)C)C)OC)C)C)O)O)/C=N/N5CCN(CC5)C)\C |
Canonical SMILES |
CC1C=CC=C(C(=O)NC2=C(C(=C3C(=C2O)C(=C(C4=C3C(=O)C(O4)(OC=CC(C(C(C(C(C(C1O)C)O)C)OC(=O)C)C)OC)C)C)O)O)C=NN5CCN(CC5)C)C |
Appearance |
Solid powder |
Color/Form |
Red to orange platelets from acetone Red-brown crystalline powde |
melting_point |
Decomposes 183-188 °C |
Purity |
>98% (or refer to the Certificate of Analysis) |
shelf_life |
>2 years if stored properly |
solubility |
Freely sol in methyl chloride, dimethyl sulfoxide; sol in tetrahydrofuran; slightly sol in water (pH less than 6), acetone, carbon tetrachloride Freely soluble in chloroform, soluble in ethyl acetate and in methanol. In water, 1,400 mg/L at 25 °C |
storage |
Dry, dark and at 0 - 4 C for short term (days to weeks) or -20 C for long term (months to years). |
Synonyms |
Rifampicin; Rifadin; Rifampin; Rimactane; Rimactan; Tubocin; Archidyn; Arficin; Arzide; Benemicin; Doloresum; Eremfat; Fenampicin; Sinerdol; |
vapor_pressure |
3.1X10-34 mm Hg at 25 °C /Estimated/ |
Origin of Product |
United States |
Historical and Foundational Research of Rifampicin
Discovery and Isolation from Microbial Sources
The journey of rifampicin began with the isolation of novel antibiotic-producing microorganisms from soil samples. This led to the identification of the rifamycin complex, a group of related compounds exhibiting antimicrobial activity.
Identification of Amycolatopsis mediterranei (formerly Nocardia mediterranei) and Related Species
The initial discovery of the rifamycins in 1957 stemmed from a screening program conducted by the Italian drug company Group Lepetit SpA in Milan. Soil samples collected near St Raphael in southern France yielded a new species of microorganism. wikipedia.orgijrpc.com This bacterium was initially classified under the genus Streptomyces and named Streptomyces mediterranei. wikipedia.orgoup.com
Later taxonomic studies led to the reclassification of this microorganism. In 1969, it was renamed Nocardia mediterranei based on its cell wall characteristics. wikipedia.orgbionity.com Further analysis in 1986, which revealed the absence of mycolic acid in its cell wall and its inability to be infected by Nocardia and Rhodococcus phages, resulted in its reclassification into a new genus, Amycolatopsis, as Amycolatopsis mediterranei. wikipedia.orgbionity.comnih.gov In 2004, based on 16S ribosomal RNA sequences, the species was renamed Amycolatopsis rifamycinica. wikipedia.org This bacterium is the primary natural producer of the rifamycin class of antibiotics. wikipedia.orgbionity.comwikipedia.orgmicrobiologyresearch.org While Amycolatopsis mediterranei is the most studied producer, other microbial sources, including marine bacteria like Salinispora, have also been found to produce rifamycin-like compounds. uq.edu.au
Initial Characterization of Rifamycin Precursors (e.g., Rifamycin B, Rifamycin SV)
The crude material extracted from the fermentation broths of Nocardia mediterranei (now Amycolatopsis mediterranei) contained a complex mixture of several microbiologically active components. oup.compsu.edu These components were initially designated rifamycin A, B, C, D, and E based on their mobility on paper chromatography. psu.edu
Isolation and characterization of these individual components proved challenging due to their instability. psu.edu The only component that could be isolated in a pure crystalline form was rifamycin B, which constituted a minor portion (5-10%) of the mixture. psu.edu Although rifamycin B itself exhibited poor antimicrobial activity, it was discovered to undergo spontaneous "activation" in aqueous oxygenated solutions. psu.edu This activation process involved the reversible conversion of rifamycin B to rifamycin O, which was then hydrolyzed to rifamycin S with the loss of a glycolic acid molecule. psu.eduasm.org Mild reduction of rifamycin S yielded rifamycin SV. psu.eduasm.org
It was subsequently demonstrated that the observed activity associated with rifamycin B was largely due to its partial transformation into these more active products, particularly rifamycin S and rifamycin SV. psu.eduasm.org Rifamycin SV became the first member of this class to be used clinically, initially administered intravenously. wikipedia.orgpsu.eduasm.org
Here is a simplified representation of the conversion pathway:
| Compound | Description | Activity Level (Relative) |
| Rifamycin B | Isolated from fermentation broth | Poorly active |
| Rifamycin O | Intermediate, formed by oxidation of Rifamycin B | - |
| Rifamycin S | Formed by hydrolysis of Rifamycin O | Highly active |
| Rifamycin SV | Formed by reduction of Rifamycin S | Highly active |
Early Chemical Modifications and Semi-Synthetic Development
Recognizing the potential of the rifamycin scaffold and the limitations of the naturally occurring compounds (such as the poor oral absorption of rifamycin SV), extensive chemical modification programs were initiated. psu.eduasm.org These efforts aimed to develop derivatives with improved pharmacological properties, including better oral bioavailability and enhanced activity against a broader spectrum of bacteria. psu.eduasm.org
Elucidation of Structure-Activity Relationships in Initial Rifamycins
Early research focused on understanding how the chemical structure of rifamycins related to their biological activity. These structure-activity relationship (SAR) studies were crucial for guiding the synthesis of new derivatives.
Initial modifications explored various functional groups on the rifamycin molecule. Changes in the ansa chain, the aliphatic bridge spanning the aromatic nucleus, generally resulted in rifamycins with significantly reduced activity compared to rifamycin SV. psu.edu Specifically, modifications involving the substitution or elimination of the two hydroxyl groups at positions C-21 and C-23 led to substantial decreases in activity. psu.eduasm.org
Most successful early SAR studies concentrated on modifications at the C-3 position of rifamycin S or SV. asm.org It was observed that electronegative groups at C-3 tended to enhance the activity, while electron-donating groups reduced activity against bacterial RNA polymerase, the molecular target of rifamycins. asm.org Modifications at the C-4 position also impacted activity, particularly concerning bacterial cell wall penetration. asm.org Rifamycins with free carboxyl groups at C-3 or C-4 showed reduced ability to penetrate the bacterial cell wall, resulting in decreased activity. asm.org
Semisynthesis Pathways Leading to this compound
The understanding gained from SAR studies fueled the development of semisynthetic rifamycin derivatives. The goal was to create compounds with improved therapeutic profiles compared to the naturally occurring rifamycins. psu.eduasm.org
This compound was developed through an extensive program of chemical modification of the rifamycins, particularly utilizing rifamycin B as a starting material despite its low intrinsic activity. oup.compsu.edunih.gov A key development was the synthesis of hydrazones of 3-formylrifamycin SV. oup.compsu.edunih.gov Among the many semisynthetic derivatives prepared, the hydrazone with N-amino-N'-methylpiperazine proved to be the most active in animal models for orally administered infections. oup.compsu.edunih.gov This compound was named this compound. ijrpc.comwikipedia.org
The development of this compound in 1965 marked a significant achievement, leading to its marketing in Italy in 1968 and subsequent approval in other countries. wikipedia.orgmicrobiologyresearch.org
Here is a simplified representation of the semisynthesis leading to this compound:
| Starting Material | Key Chemical Transformation | Product |
| Rifamycin SV | Formation of 3-formylrifamycin SV, then reaction with N-amino-N'-methylpiperazine | This compound |
Molecular Mechanisms of Action of Rifampicin
Inhibition of Bacterial RNA Polymerase (RNAP)
The primary mechanism by which rifampicin inhibits bacterial growth is through the potent inhibition of bacterial RNAP. asm.orgdroracle.ainih.gov This enzyme is responsible for synthesizing RNA from a DNA template. By binding to RNAP, this compound effectively halts RNA synthesis, preventing the production of essential proteins and leading to bacterial cell death. droracle.aipatsnap.com
Specific Binding Site Characterization on the RNAP β-Subunit (RpoB)
This compound achieves its inhibitory effect by binding to a specific pocket within the bacterial RNAP. This binding site is predominantly located on the β-subunit of the enzyme, encoded by the rpoB gene. asm.orgpatsnap.comnih.govrichmond.eduwikipedia.org The high-affinity binding of this compound to this site is crucial for its antibacterial potency. asm.orgpnas.org Mutations in the rpoB gene are frequently associated with this compound resistance, further highlighting the importance of this binding site. asm.orgnih.govrichmond.eduwikipedia.org
Structural studies, particularly X-ray crystallography and cryo-EM, have provided detailed insights into the interaction between this compound and bacterial RNAP. asm.orgnih.govmdpi.com These studies have shown that this compound binds within the DNA/RNA binding channel of the RNAP β-subunit. asm.orgnih.govnih.govoup.com The binding pocket is located in proximity to the enzyme's active site, typically within 12 to 20 Å of the catalytic Mg²⁺ ion. asm.orgnih.govkoreamed.org The structural data support a model where the bound this compound molecule physically obstructs the path of the nascent RNA chain during transcription initiation. nih.govkoreamed.orgpnas.orgpnas.org
Crystal structures of RNAP from organisms like Thermus aquaticus and Escherichia coli in complex with this compound have been instrumental in visualizing these interactions. asm.orgnih.govnih.govrichmond.edumdpi.comkoreamed.org These structures reveal the precise location of the this compound binding site and how it overlaps with the region where the newly synthesized RNA transcript emerges from the enzyme. richmond.edukoreamed.org
The binding of this compound to the β-subunit of RNAP involves interactions with several key amino acid residues within the binding pocket. asm.orgnih.govrichmond.edu These residues contribute to the binding affinity and specificity of this compound. Mutations in these specific amino acids can lead to reduced binding affinity and consequently, this compound resistance. asm.orgnih.govrichmond.eduwikipedia.orgoup.com
Studies have identified several residues within the this compound Resistance-Determining Region (RRDR) of RpoB as critical for this compound binding. nih.govrichmond.eduwikipedia.org For example, in E. coli and M. tuberculosis, mutations at positions such as Ser531, His526, and Asp516 are frequently observed in resistant strains. asm.orgnih.govwikipedia.orgasm.org These residues can form hydrogen bonds or other interactions with the this compound molecule. asm.orgnih.gov Alterations in these residues can disrupt these interactions, reducing the stability of the this compound-RNAP complex. nih.govasm.org
The following table summarizes some key amino acid residues in E. coli RpoB involved in this compound binding and the impact of mutations:
| E. coli RpoB Residue (Numbering) | Interaction with this compound | Impact of Mutation on Resistance | Source |
| Asp516 | Involved in shaping the binding pocket. | Can reduce affinity. | nih.govrichmond.eduasm.org |
| His526 | Can form interactions with this compound. | Can prevent binding due to steric conflicts. nih.gov | asm.orgnih.govrichmond.eduwikipedia.org |
| Ser531 | Forms a hydrogen bond with this compound. | Reduces affinity by disordering binding interface. nih.govasm.org | asm.orgnih.govrichmond.eduwikipedia.orgasm.org |
| Ile572 | Located near the binding site. | Mutations can confer resistance. richmond.eduasm.org | richmond.eduasm.org |
| Pro519 (S. aureus numbering, corresponds to P564 in E. coli) | Located in a conserved region near the binding site. | Can confer high-level resistance. asm.org | asm.org |
Note: Residue numbering can vary between bacterial species. E. coli numbering is commonly used as a reference. asm.orgnih.govwikipedia.org
Structural Biology Insights into this compound-RNAP Complex
Mechanism of Transcription Inhibition at the Initiation Phase
This compound primarily inhibits bacterial transcription during the initiation phase. asm.orgdroracle.aikoreamed.orgresearchgate.net It binds to RNAP before the synthesis of a stable, elongated RNA transcript. Once RNAP has synthesized an RNA chain beyond a certain length (typically 2-3 nucleotides), it becomes much less susceptible to this compound inhibition. patsnap.comnih.govpnas.orgoup.comkoreamed.orgpnas.orgpnas.orgresearchgate.net
The binding of this compound prevents the transition from an initial, short RNA product to a longer, elongating transcript. nih.govkoreamed.orgpnas.org This leads to the release of short, "abortive" RNA transcripts, usually only a few nucleotides in length, as the transcription process is prematurely halted. koreamed.orgpnas.orgnih.gov
The structural location of this compound within the RNAP active site channel explains its ability to block nascent RNA elongation. nih.govnih.govrichmond.edukoreamed.orgpnas.orgnih.gov By occupying space within this channel, the this compound molecule physically obstructs the path that the growing RNA chain would normally follow as it exits the enzyme. patsnap.comnih.govoup.comkoreamed.orgpnas.orgresearchgate.net This steric clash prevents the RNAP from synthesizing RNA transcripts longer than 2-3 nucleotides. patsnap.comnih.govpnas.orgoup.comkoreamed.orgpnas.orgpnas.orgresearchgate.net The nascent RNA chain simply cannot extend further due to the physical presence of this compound in its path. nih.govkoreamed.orgresearchgate.net
While the primary mechanism is steric hindrance of elongation, some research also suggests that this compound may interfere with the formation of the second or third phosphodiester bond during transcription initiation. asm.orgpnas.orgpnas.orgnih.gov Although this compound does not typically affect the formation of the first phosphodiester bond, its presence in the active site region can impede the subsequent steps required for extending the RNA chain. asm.orgpnas.orgkoreamed.orgpnas.org This interference is thought to be a consequence of the drug's binding position, which, while not directly at the catalytic center, is close enough to influence the precise positioning of substrates (nucleotide triphosphates) and the nascent RNA required for efficient phosphodiester bond formation beyond the initial few nucleotides. asm.orgpnas.orgnih.gov
Blockage of Nascent RNA Elongation
Cellular and Physiological Responses to RNAP Inhibition
Impact on Bacterial Gene Expression and Protein Synthesis Pathways
The primary consequence of this compound binding to RNAP is the direct inhibition of bacterial transcription. By blocking the elongation of mRNA synthesis, this compound effectively halts the production of new mRNA molecules wikipedia.orgnih.gov. Since mRNA serves as the template for protein synthesis, the cessation of transcription leads to a rapid decline in the availability of new transcripts. While existing mRNA molecules may continue to be translated for a period, their relatively short half-lives in bacteria (around 5 minutes in E. coli) mean that protein synthesis pathways are quickly impacted nih.gov. This disruption of gene expression and subsequent protein synthesis is a critical factor in the antibacterial activity of this compound wikipedia.orgmdpi.com. Studies have shown that this compound treatment prevents the synthesis of host bacterial proteins wikipedia.org.
Modulation of Bacterial Metabolic States by this compound
Inhibition of RNAP by this compound has been shown to influence various aspects of bacterial metabolism. These metabolic alterations can play a role in the drug's efficacy and the bacterial response to treatment.
Effects on Aerobic Respiration
Blocking RNA synthesis with this compound has been linked to a reduced rate of aerobic respiration in bacteria. nih.govasm.org While for most bactericidal antibiotics, the inhibition of bacterial respiration can counteract killing, research suggests that reduced aerobic respiration might contribute to this compound-induced killing in E. coli. nih.gov
Influence on DNA Repair Mechanisms
The killing efficacy of this compound can be influenced by bacterial DNA repair mechanisms. nih.govasm.org Evidence suggests that the maintenance of DNA replication and transcription-coupled nucleotide excision repair can protect E. coli cells against this compound killing. nih.govasm.org This highlights a link between the core process of transcription inhibition and the cellular pathways responsible for maintaining genomic integrity in the face of antibiotic stress.
Differentiating Bacteriostatic and Bactericidal Actions at the Molecular Level
This compound is generally considered to have bactericidal activity, meaning it kills bacteria, although its effect can be bacteriostatic (inhibiting growth) in some contexts or against certain organisms, such as E. coli. rcsb.orgnih.govplos.org The bactericidal action of this compound and other rifamycins is attributed to their binding to the beta subunit of bacterial RNA polymerase (RpoB), which blocks the elongation of the forming RNA transcript beyond a few nucleotides (2-3 nt). researchgate.net This direct inhibition of a vital process like transcription initiation is fundamentally disruptive to bacterial cellular function. nih.gov While the molecular basis for the distinction between bacteriostatic and bactericidal effects can be complex and may involve multiple factors including the bacterial species and its metabolic state, the direct and potent inhibition of RNAP at the initiation stage is the core molecular event underlying this compound's killing potential. nih.govplos.orgresearchgate.net
Molecular Mechanisms of Rifampicin Resistance
Target-Based Resistance: Mutations in the rpoB Gene
Mutations in the rpoB gene are the most common cause of rifampicin resistance in various bacteria, including Mycobacterium tuberculosis, Escherichia coli, and Staphylococcus aureus. mcmaster.cafrontiersin.orgasm.org These mutations typically result in amino acid substitutions in the RpoB protein that interfere with this compound binding. mcmaster.ca
Characterization of the this compound Resistance-Determining Region (RRDR)
A specific region within the rpoB gene, known as the this compound Resistance-Determining Region (RRDR), is a hotspot for resistance-conferring mutations. tandfonline.comnih.govasm.orgnih.gov In Mycobacterium tuberculosis, this region spans codons 426 to 452, which corresponds to codons 507 to 533 in the Escherichia coli numbering scheme. tandfonline.comnih.govnih.gov The vast majority, approximately 90-96%, of this compound-resistant clinical isolates of M. tuberculosis harbor mutations within this 81-base pair region. nih.govnih.govnih.govbesjournal.comnih.gov
Identification of Specific Amino Acid Substitutions Conferring Resistance
Amino acid substitutions within the RRDR are primarily responsible for conferring this compound resistance. asm.orgnih.govnih.govnih.govnih.gov The specific substitution and its location within the RRDR can influence the level of resistance.
High-Level Resistance Mutations (e.g., S531, H526)
Certain amino acid substitutions frequently confer high levels of resistance to this compound. In E. coli and M. tuberculosis, mutations at positions homologous to E. coli RpoB codons D516, H526, and S531 are commonly associated with high-level resistance in clinical isolates. frontiersin.orgoup.com For instance, the S531L mutation in M. tuberculosis is frequently observed and is linked to high this compound resistance. nih.govoup.combrieflands.comfrontiersin.org Similarly, mutations at H526 (such as H526Y and H526D) are also prevalent high-level resistance mutations. frontiersin.orgnih.govoup.combrieflands.comfrontiersin.org Studies have shown that these specific mutations can individually result in a high minimum inhibitory concentration (MIC) of this compound. oup.comoup.com
Data Table: Common High-Level this compound Resistance Mutations in M. tuberculosis (based on E. coli numbering)
| E. coli RpoB Codon | M. tuberculosis RpoB Codon (approximate) | Common Substitutions | Resistance Level | Frequency in Clinical Isolates |
| D516 | D435, D441 | V, Y, G, N | High/Moderate | Varies, D435V associated with moderate resistance nih.govresearchgate.net, D516V frequent in some regions nih.govfrontiersin.org |
| H526 | H445, H451 | Y, D, R, N, L | High | Frequent frontiersin.orgnih.govoup.combrieflands.comfrontiersin.org, H445D/Y/R associated with high resistance nih.govdovepress.com, H451D/Y/R linked to altered binding mdpi.com |
| S531 | S450, S456 | L, F | High | Most frequent nih.govoup.combrieflands.comfrontiersin.org, S450L associated with high resistance nih.govdovepress.com |
Low-Level Resistance Mutations (e.g., H451D/Y/R, mutations outside RRDR)
While the RRDR is the primary site for resistance mutations, mutations outside this region can also confer this compound resistance, although often at lower levels. mcmaster.cadovepress.comnih.govmdpi.com Some mutations within the RRDR may also result in lower levels of resistance compared to the common high-level mutations. oup.comoup.com For example, mutations at M. tuberculosis codon H451 (corresponding to E. coli H526) with substitutions like H451D, H451Y, or H451R have been studied for their impact on this compound binding and resistance. mdpi.commdpi.com Mutations outside the RRDR, such as Ile491Phe in M. tuberculosis, have been identified and can confer resistance that may not be detected by standard RRDR-based molecular tests. dovepress.comnih.gov Low-level resistance mutations within the RRDR are sometimes found in conjunction with other rpoB mutations. oup.comoup.com
Impact of rpoB Mutations on RNAP-Rifampicin Affinity
Mutations in the rpoB gene, particularly within the RRDR, primarily confer resistance by decreasing the affinity of RNAP for this compound. nih.govmcmaster.cafrontiersin.orgtandfonline.comasm.orgnih.gov These amino acid substitutions alter the structure of the this compound binding pocket on the β-subunit of RNAP, hindering the drug's ability to bind effectively. nih.govtandfonline.comnih.govresearchgate.net Structural studies have shown that mutations like S531L can subtly impact the binding interface, while mutations such as H526Y can reshape the binding pocket, creating steric clashes that prevent this compound binding. researchgate.net Even mutations outside the RRDR can influence this compound binding efficiency by causing structural changes in RpoB. mdpi.com
Non-Target-Based Resistance Mechanisms
Beyond modifications to the drug's primary target, bacterial cells can employ mechanisms that reduce the intracellular concentration of this compound or chemically alter the drug, rendering it inactive. acs.orgfrontiersin.org These non-target mechanisms are crucial aspects of the bacterial resistome against rifamycins. sci-hub.senih.gov
Efflux Pump Systems
Bacterial efflux pumps are membrane proteins that actively transport various substrates, including antibiotics, out of the cell. asm.orgtandfonline.com These systems can contribute to antibiotic resistance by reducing the intracellular accumulation of the drug below inhibitory concentrations. asm.org
Identification of Specific Efflux Pump Genes (e.g., Rv0783, Rv2936, Rv0933)
Studies investigating this compound resistance in M. tuberculosis isolates lacking mutations in rpoB have identified the overexpression of specific efflux pump genes. Transcriptional analyses have indicated that genes such as Rv0783, Rv2936, and Rv0933 are overexpressed in some this compound-resistant clinical isolates. mdpi.comnih.govoup.com These findings suggest a potential role for these efflux pumps in the observed resistance phenotype. mdpi.comnih.gov Another efflux pump gene, Rv1258c (Tap), has also been investigated, with a novel mutation identified in a this compound mono-resistant clinical strain. jidc.org
Enzymatic Modification of this compound
Bacteria can also inactivate this compound through enzymatic modifications that alter the drug's structure, preventing its effective binding to RNAP. acs.orgsci-hub.senih.gov
ADP Ribosylation
ADP ribosylation is an enzymatic process where an ADP-ribose moiety is transferred from NAD+ to an acceptor molecule. While primarily known as a post-translational modification of proteins, rifamycin antibiotics are the only known small-molecule targets for ADP ribosylation-mediated inactivation. acs.orgbiorxiv.org This modification is catalyzed by enzymes known as this compound ADP-ribosyltransferases (Arr). acs.orgnih.govasm.org The Arr enzyme transfers the ADP-ribose group to the hydroxyl group at the C23 position of this compound. acs.orgsci-hub.seacs.orgbiorxiv.orgnih.gov This modification at a critical position for interaction with RNAP sterically blocks the productive binding of this compound to the enzyme, thereby inactivating the antibiotic. acs.orgsci-hub.seasm.org Arr-mediated this compound resistance was first identified in Mycolicibacterium smegmatis. acs.orgbiorxiv.orgnih.gov Arr homologues are found in a variety of bacterial species, including pathogens, and are often associated with mobile genetic elements. acs.orgbiorxiv.orgnih.gov
Rifamycin Monooxygenase (Rox) Activity and Degradation Pathways
Another enzymatic mechanism involves rifamycin monooxygenases (Rox), which are FAD-dependent enzymes. acs.orgnih.govfrontiersin.org Rox enzymes catalyze the hydroxylation of the naphthoquinone core of rifamycins. acs.orgnih.gov This hydroxylation initiates a degradation pathway that leads to the untethering of the ansa chain from the naphthyl moiety. acs.orgsci-hub.senih.gov This structural disruption prevents the antibiotic from adopting the necessary 3D conformation required for binding to the RNA exit tunnel of RNAP, thus providing resistance. sci-hub.senih.govmcmaster.carcsb.org Rox-catalyzed rifamycin degradation has been characterized in various environmental bacteria and is associated with the decomposition of this compound. frontiersin.orgnih.govmcmaster.carcsb.org The rox gene was initially identified in Rhodococcus equi. frontiersin.orgfrontiersin.org Studies have shown that Rox proteins from rifamycin producers also possess enzymatic activity against rifamycins like this compound. frontiersin.orgfrontiersin.org
Interactive Data Table 2: Enzymatic Modifications of this compound Conferring Resistance
| Modification Type | Enzyme Class | Modification Site on this compound | Mechanism of Inactivation |
| ADP Ribosylation | ADP-ribosyltransferases (Arr) | C23 hydroxyl group acs.orgsci-hub.senih.gov | Sterically blocks binding to RNAP acs.orgsci-hub.seasm.org |
| Hydroxylation/Degradation | Rifamycin monooxygenases (Rox) | Naphthoquinone core acs.orgnih.gov | Leads to structural disruption, preventing RNAP binding sci-hub.senih.govmcmaster.carcsb.org |
| Phosphorylation | Rifampin phosphotransferases (RPH) | C21 hydroxyl group pnas.org | Leads to steric clash, weakening or abolishing binding to RNAP pnas.org (Mentioned in sources but outside strict outline scope) |
| Glycosylation | Glycosyltransferases | C23 hydroxyl group acs.orgsci-hub.se | Modifies the hydroxyl group, blocking interaction with RNAP acs.orgsci-hub.se (Mentioned in sources but outside strict outline scope) |
3.2.3. Alterations in Bacterial Cell Wall Permeability
Alterations in bacterial cell wall permeability represent a significant mechanism contributing to this compound resistance, particularly in organisms like Mycobacterium tuberculosis and other mycobacteria, which possess a uniquely complex and lipid-rich cell wall structure oup.comfrontiersin.orgasm.org. This intrinsic barrier can restrict the influx of antibiotics, including this compound, thereby contributing to natural or acquired resistance oup.comfrontiersin.orgasm.orgmdpi.com.
Studies have proposed that impaired permeability to this compound is a primary cause of natural resistance in some mycobacteria, such as Mycobacterium smegmatis, Mycobacterium avium, and Mycobacterium intracellulare, even when their RNA polymerase molecules are susceptible to the drug oup.com. The uptake of this compound, a hydrophobic antibiotic, is generally thought to occur via passive diffusion across the lipid-rich mycobacterial cell wall oup.comfrontiersin.org. Reduced rates of drug influx due to changes in this barrier can therefore lead to resistance oup.com.
Research into this compound-surviving populations of Mycobacterium tuberculosis has revealed the development of a thickened capsular outer layer (TOL) frontiersin.orgbiorxiv.orgresearchgate.net. This thickened layer, characterized by a high abundance of specific polysaccharides including α-D-glucopyranoside, 1,2,5-linked-mannitol, and 3,4-linked mannose, appears to function as a physical barrier that restricts the permeability of this compound into the bacterial cells frontiersin.orgbiorxiv.orgresearchgate.net. The increased thickness and elevated negative charge of this layer are hypothesized to impede the entry of the relatively non-polar this compound molecule frontiersin.org. Mechanical removal of this thickened outer layer has been shown to significantly enhance the permeability of fluorochrome-conjugated this compound into persistent cells, further supporting its role as a barrier biorxiv.orgresearchgate.net.
While the primary mechanism of this compound resistance often involves mutations in the rpoB gene encoding the RNA polymerase β-subunit, alterations in cell wall permeability can act as a complementary mechanism or contribute to intrinsic resistance oup.comnih.govresearchgate.net. In some cases, this compound resistance has been observed in strains without rpoB mutations, suggesting the involvement of alternative mechanisms like permeability barriers mdpi.com.
Changes in cell wall composition beyond thickening can also play a role. For instance, this compound resistance mutations have been associated with chemical remodeling of the Mycobacterium tuberculosis cell wall, including altered concentrations of cell surface polyketide lipids nih.gov. These lipids are known to control the permeability of the organism and could affect the transport of drugs nih.gov.
Although efflux pumps are distinct mechanisms, their activity can synergize with permeability barriers to reduce the intracellular concentration of antimicrobials frontiersin.orgasm.org. However, studies specifically investigating the role of the thickened outer layer in this compound persistence in M. tuberculosis found that efflux pump inhibitors did not affect the intracellular levels of fluorochrome-conjugated this compound, suggesting that in this specific context, the reduced permeability was primarily due to the altered cell wall structure rather than increased efflux activity biorxiv.orgresearchgate.net.
In Gram-negative bacteria, porins in the outer membrane play a crucial role in the passive transport of hydrophilic compounds, including some antibiotics nih.govfrontiersin.orgnih.gov. While this compound is relatively hydrophobic and thought to primarily diffuse across membranes, alterations in porin expression or function have been linked to resistance to various antibiotics, and some studies in Escherichia coli have indicated that specific porin mutations can affect this compound resistance nih.govfrontiersin.orgbiorxiv.org. However, the impact of porin alterations on this compound permeability appears to be less prominent compared to their role in the transport of more hydrophilic drugs nih.govfrontiersin.orgnih.gov.
Biosynthesis and Semi-synthetic Pathways of Rifampicin
Natural Product Biosynthesis of Rifamycins
The biosynthesis of rifamycins, the parent compounds of rifampicin, is a complex process carried out by the bacterium Amycolatopsis mediterranei. This process involves a type I polyketide synthase (PKS) system and a series of subsequent modification steps. The resulting natural product, typically rifamycin B, serves as a crucial precursor for the semi-synthetic production of this compound. nih.govnih.gov
Polyketide Synthase (PKS) Machinery in Amycolatopsis mediterranei
The core structure of rifamycins is assembled by a type I polyketide synthase (PKS) machinery encoded by the rifA through rifE genes in A. mediterranei. nih.govpnas.orgwikipedia.orgnih.gov This modular PKS system is responsible for the iterative condensation of specific precursor molecules, building the large macrocyclic ansa chain characteristic of rifamycins. nih.govpnas.orgwikipedia.org The PKS operates in a processive manner, assembling the polyketide chain. pnas.orgnih.gov
Precursor Incorporation and Chain Elongation
The biosynthesis of the rifamycin carbon skeleton begins with an unusual starter unit, 3-amino-5-hydroxybenzoic acid (AHBA). nih.govpnas.orgwikipedia.orgbionity.com AHBA is believed to originate from the shikimate pathway, although its direct incorporation mechanism has been studied using labeled precursors. wikipedia.orgbionity.com Following the initiation with AHBA, the PKS machinery incorporates extender units, specifically two acetate and eight propionate units, through malonyl-CoA and methylmalonyl-CoA, respectively. nih.govwikipedia.orgbionity.comchimia.ch These units are sequentially added to the growing polyketide chain on the PKS assembly line. nih.gov
The incorporation pattern of these precursors into rifamycin S has been established through isotopic labeling experiments. chimia.ch
| Precursor | Number of Units Incorporated |
| 3-amino-5-hydroxybenzoic acid (AHBA) | 1 |
| Acetate | 2 |
| Propionate | 8 |
Post-PKS Modification and Cyclization Reactions
Once the linear undecaketide chain is assembled by the PKS, it undergoes a series of post-PKS modifications and cyclization reactions to form the mature rifamycin structure. nih.govresearchgate.net The rifF gene product, an amide synthase, is responsible for the release of the completed linear polyketide from the PKS and the formation of an intramolecular amide bond, leading to a macrocyclic lactam structure, presumably proansamycin X. pnas.orgnih.govnih.govmicrobiologyresearch.org
Oxidative cyclization to form the naphthoquinone chromophore occurs during PKS assembly, rather than after. pnas.orgnih.gov This process involves the formation of an 8-hydroxy-7,8-dihydronaphthoquinone structure, which is subsequently dehydrogenated to the 8-hydroxynaphthoquinone found in mature rifamycins like rifamycin B. pnas.orgnih.gov Further tailoring modifications, such as hydroxylation, methylation, and acetylation, contribute to the structural diversity of natural rifamycins. rsc.org Rifamycin W is an intermediate that undergoes rearrangement and oxidative cleavage to produce rifamycin B. researchgate.net
Chemical and Semi-Synthetic Strategies for this compound Production
This compound is a semi-synthetic derivative, meaning it is not directly produced by fermentation but is synthesized chemically from a naturally produced rifamycin, primarily rifamycin S or rifamycin SV. acs.orgnih.govbasicmedicalkey.commsdvetmanual.com This approach leverages the microbial machinery for the complex polyketide synthesis and utilizes efficient chemical reactions for the final structural modifications.
Conversion of Rifamycin S to this compound via Cyclization and Condensation
The primary semi-synthetic route to this compound involves the conversion of rifamycin S. researchgate.netgoogle.comgoogle.com Rifamycin S is typically obtained by oxidation of rifamycin B, the main fermentation product of A. mediterranei. chimia.chacs.orgbasicmedicalkey.com Rifamycin S can also be prepared from rifamycin SV. acs.org
The synthesis of this compound from rifamycin S generally involves a cyclization reaction followed by a condensation reaction. researchgate.net One common method involves reacting rifamycin S with a 1,3,5-trisubstituted hexahydro-1,3,5-triazine (such as 1,3,5-trimethyl-hexahydro-1,3,5-triazine) in an aprotic dipolar solvent, optionally in the presence of formaldehyde. nih.govgoogle.comgoogle.com This step leads to the formation of an intermediate, often a 1,3-oxazino rifamycin derivative. nih.govgoogle.comgoogleapis.com
Subsequently, this intermediate undergoes a condensation reaction with 1-amino-4-methylpiperazine to yield this compound. nih.govresearchgate.netgoogle.comgoogle.comgoogleapis.com This reaction is typically carried out while maintaining the pH in a specific range, often between 5 and 7. google.comgoogle.com
A simplified representation of the key chemical transformation is the reaction of 3-formylrifamycin SV (derived from rifamycin SV) with 1-amino-4-methylpiperazine, forming a hydrazone (this compound). basicmedicalkey.compsu.edu
Optimization of Reaction Conditions and Yields
Optimization of the semi-synthetic process is crucial for improving the yield and purity of this compound. Research has focused on various aspects, including reaction temperatures, reaction times, solvent systems, and the ratios of reactants and catalysts. researchgate.netgoogle.comgoogle.com
For the cyclization reaction, conditions such as a temperature of 55°C and a reaction time of 1.5 hours have been investigated. researchgate.net The ratio of reactants, such as N,N-dihydroxymethyl tert-butylamine, glacial acetic acid, ascorbic acid, and rifamycin S, has been optimized to improve yield and purity. researchgate.net
For the subsequent condensation reaction with 1-amino-4-methylpiperazine, reaction time and temperature, as well as the ratio of 1-amino-4-methylpiperazine to rifamycin S, are critical parameters. researchgate.net Maintaining the pH during the condensation step is also important for optimal results. google.comgoogle.com
Chemical Derivatization of the Rifamycin Core
The rifamycins, characterized by their ansa structure consisting of a naphthalene core bridged by an aliphatic chain, serve as the foundational scaffold for the synthesis of numerous semi-synthetic derivatives, including this compound. These modifications are primarily undertaken to enhance pharmacological properties such as bioavailability, spectrum of activity, and to overcome resistance mechanisms nih.govfrontiersin.orgrsc.org. The chemical derivatization process typically begins with naturally occurring rifamycins, such as rifamycin B, which is produced by Amycolatopsis rifamycinica (formerly Nocardia mediterranei) wikipedia.orggoogle.commdpi.com.
Rifamycin B itself has limited antibiotic activity and is often converted to more active forms like rifamycin O and rifamycin S through chemical oxidation and hydrolysis google.commdpi.com. Rifamycin S is a key intermediate for the synthesis of many rifamycin derivatives google.commdpi.com.
Historically, the most significant chemical modifications have been focused on the C-3 and C-4 positions of the rifamycin naphthalene ring system nih.govfrontiersin.org. These modifications have been crucial in developing clinically important drugs with improved properties compared to the parent compounds frontiersin.orgfrontiersin.org. For instance, this compound is a semi-synthetic derivative obtained from rifamycin B, chemically derived via rifamycin S and rifamycin SV mdpi.com. Specifically, this compound is described as the hydrazone of a rifamycin B derivative with N-amino-N'-methylpiperazine mdpi.com. The synthesis of this compound from 24-desmethylrifamycin S, for example, involves treatment with paraformaldehyde and 1,3,5-trimethyl-hexahydro-1,3,5-triazine, followed by reaction with 1-amino-4-methylpiperazine nih.gov.
Other clinically used rifamycin derivatives, such as Rifabutin and Rifapentine, are also products of semi-synthetic modifications of the rifamycin core wikipedia.orgwikipedia.orgwikipedia.orgrcsb.org. Rifabutin is a derivative of rifamycin S wikipedia.org. Rifapentine, synthesized from this compound, involves the substitution of a methyl group with a cyclopentane group wikipedia.org.
While modifications at the C-3 and C-4 positions have been extensively explored and have yielded the majority of clinically used rifamycins, research continues into modifying other positions on the rifamycin core to generate novel analogs with improved activity, particularly against resistant strains. For example, modifications at the C-25 position of the rifamycin ansa-chain have been investigated as a strategy to circumvent resistance mediated by ADP-ribosylation oup.comnih.govd-nb.info. Studies have shown that introducing various carbamates or other groups at the C-25 position can prevent enzymatic inactivation while maintaining activity against bacterial RNA polymerase oup.comnih.govd-nb.info.
Structure-activity Relationship Sar Studies of Rifampicin and Its Analogs
Identification of Key Pharmacophoric Features
The pharmacophore of a molecule represents the ensemble of steric and electronic features necessary to ensure optimal supramolecular interactions with a specific biological target structure, triggering or blocking its biological response. For rifampicin, the interaction with bacterial RNAP is key to its activity.
Importance of Macrocyclic Ring and Ansa Chain
This compound belongs to the ansamycin family of antibiotics, characterized by a macrocyclic ring structure (the ansa chain) that bridges two non-adjacent positions of an aromatic moiety (the naphthoquinone core). This macrocyclic structure is fundamental to the activity of rifamycins. The ansa chain, a polyketide-derived fragment, plays a crucial role in positioning the molecule within the RNAP binding pocket and facilitating key interactions. ubc.ca The flexibility of the ansa chain is also considered important for the biological activity of rifamycins, potentially allowing for adaptation to the cellular environment and the target site. rsc.orgresearchgate.net Studies on rifamycin derivatives with more rigid ansa chains have shown lower antibacterial potency, highlighting the importance of this flexibility. researchgate.netresearchgate.net
Influence of Stereochemistry and Conformational Flexibility on Activity
The stereochemistry (the three-dimensional arrangement of atoms) and conformational flexibility (the ability of the molecule to adopt different shapes) of this compound are critical determinants of its ability to bind effectively to bacterial RNAP.
Analysis of Zwitterionic and Pseudocyclic Structures
Rifamycin congeners can exist in solution in different structural forms, including zwitterionic and pseudocyclic structures. researchgate.netresearchgate.netnih.gov The zwitterionic form can arise from proton transfer, for example, from a phenolic hydroxyl group to a basic nitrogen atom. researchgate.netresearchgate.netnih.govrsc.org Pseudocyclic structures can be stabilized by intramolecular hydrogen bonds. researchgate.netnih.gov The presence and distribution of these different forms in solution can influence the molecule's physicochemical properties, such as lipophilicity and solubility, which in turn affect its antibacterial properties. researchgate.netresearchgate.netnih.gov Spectroscopic studies, such as NMR, are used to investigate these structural forms in solution. researchgate.netresearchgate.netresearchgate.netnih.govdiva-portal.org
Rational Design and Synthesis of Novel Rifamycin Derivatives
Understanding the SAR of this compound provides a basis for the rational design and synthesis of novel rifamycin derivatives with improved properties. By identifying key structural features and their contributions to activity, researchers can make targeted modifications to the this compound scaffold to enhance potency, broaden the spectrum of activity, improve pharmacokinetic profiles, or overcome resistance mechanisms. researchgate.netnih.gov For example, modifications at the C(3) position have been explored to yield derivatives with altered antibacterial activity and physicochemical properties. researchgate.netresearchgate.netnih.gov The synthesis of novel rifamycin derivatives often involves chemical modifications of naturally occurring rifamycins or their precursors. researchgate.netrsc.org Computational approaches, such as molecular docking and dynamics simulations, are valuable tools in the rational design process, allowing researchers to predict the binding modes and affinities of potential new derivatives and guide synthetic efforts. frontiersin.orgcabidigitallibrary.org The goal is to create new compounds that maintain or improve the desirable characteristics of this compound while addressing limitations such as resistance development. researchgate.netrsc.orgnih.gov
Targeted Modifications to Improve Potency and Spectrum
Modifications to the rifamycin scaffold have been explored to enhance antibacterial activity and broaden the spectrum. Historically, synthetic modifications at the C-3/C-4 region of the rifamycin naphthalene core have yielded significant improvements in pharmacological properties. acs.org this compound itself contains a methylpiperazine group appended to the C-3 position. acs.org
Analogs with improved activity against various Mycobacterium species and other bacteria have been developed. For instance, some rifamycin derivatives, such as CGP 7040, have demonstrated lower minimum inhibitory concentrations (MICs) compared to this compound against sensitive M. tuberculosis strains. sciepub.com CGP7040 also showed superior activity against M. avium and improved stability relative to this compound. sciepub.com
Further research has focused on modifying the rifamycin polyketide backbone. For example, 24-desmethylrifamycin, generated through a combined genetic-synthetic strategy, exhibited better antibacterial activity than this compound against multidrug-resistant strains of Mycobacterium tuberculosis. researchgate.netnih.gov This suggests that modifications beyond the traditional C-3/C-4 positions can lead to improved analogs.
Studies involving aminomethyl rifamycin SV derivatives have explored appending aminoalkyl-aromatic ring tails to the C3 position. ucl.ac.uk Replacing the typical hydrazone unit of this compound with an amino-alkyl linkage connecting the aromatic ring tails to the rifamycin naphthoquinone core resulted in novel analogs. ucl.ac.uk Some of these analogs showed anti-tubercular activity against wild-type and certain this compound-resistant strains, with some exhibiting activity comparable to or higher than this compound against specific mutated strains. ucl.ac.uk In silico analysis suggested a distinct binding mode for these new analogs within the RNAP binding pocket compared to this compound. ucl.ac.uk
Data on the comparative activities of some rifamycin analogs against Mycobacterium avium infection in a mouse model indicated that rifabutin and rifapentine had comparable activities at a specific dosage, while other analogs like P/DEA and CGP 7040 were less active at the same dose. nih.gov
Strategies to Overcome Resistance (e.g., modifications to circumvent rpoB mutations or efflux)
This compound resistance primarily arises from mutations in the rpoB gene, leading to amino acid substitutions in the RNAP β-subunit, particularly within the this compound resistance-determining region (RRDR). tandfonline.comontosight.aimdpi.comresearchgate.netnih.gov These mutations alter the this compound binding site, reducing the drug's affinity for the enzyme. tandfonline.comresearchgate.net Resistance can also involve efflux pumps that actively remove the drug from bacterial cells. ontosight.airesearchgate.net
Developing this compound analogs that can overcome these resistance mechanisms is a critical area of research. Modifications of the C3 tails of this compound have shown potential in leading to analogs with enhanced antibiotic activity against this compound-resistant M. tuberculosis strains bearing mutations in the RRDR sequence. tandfonline.com
Some newer rifamycin analogs, such as certain C-3/C-4 benzoxazino Kanglemycin derivatives, have demonstrated improved activity against wild-type bacteria and acquired activity against clinically relevant this compound-resistant mutations, including the S456L mutation in M. tuberculosis RNAP. acs.org While the parent compound, Kanglemycin A, showed activity against the S486L mutant in S. aureus (corresponding to M. tuberculosis S456L), it was not active against the H481Y variant (corresponding to M. tuberculosis H451Y), whereas this compound was inactive against both mutants. acs.org Certain C-3/C-4 derivatives showed significant improvements in activity against the this compound-resistant S456L strain compared to Kanglemycin A. acs.org
The improved activity of some analogs against this compound-resistant strains suggests that further work, particularly utilizing information from co-crystal structures with RNAP, could lead to even better analogs capable of circumventing resistance. researchgate.net
Efflux pumps are another mechanism of resistance in bacteria, including mycobacteria, that can reduce the intracellular concentration of this compound. ontosight.airesearchgate.netmdpi.com While some studies have suggested that the addition of efflux pump inhibitors did not significantly modulate the intracellular accumulation of this compound in drug-susceptible mycobacteria, targeting efflux systems remains a potential strategy to enhance the efficacy of anti-TB therapy, potentially in combination with this compound or its analogs. oup.com
Newer drugs and analogs are being developed with the aim of being effective against multidrug and extensively drug-resistant strains. sciepub.com For example, bedaquiline (BDQ), a diarylquinoline, has a different mechanism of action, inhibiting ATP synthesis, and has been approved for the treatment of multidrug-resistant tuberculosis. wikipedia.orgescholarship.orgresearchgate.net While not a this compound analog, its development highlights the need for drugs with novel mechanisms to combat resistance. Other compounds like PBTZ169, a benzothiazinone derivative, target DprE1, an essential enzyme in cell wall biosynthesis, and have shown nanomolar bactericidal activity against M. tuberculosis in vitro. invivochem.comuni.lunih.govresearchgate.netcuni.cz
The following table summarizes some of the compounds mentioned and their PubChem CIDs:
| Compound Name | PubChem CID |
| This compound | 135398735 rcsb.orgmims.comnih.gov |
| Rifabutin | 6323490 wikipedia.orgnih.govnih.govescholarship.org |
| Rifapentine | 6323497 escholarship.orgwikipedia.orgresearchgate.netmims.comfishersci.beuni.lu |
| Rifaximin | 6436173 escholarship.orgwikipedia.orgwikidata.orgmims.comzellbio.eu |
| KRM-1648 | 135431094 idrblab.net (also known as Rifalazil escholarship.orgwikipedia.org) |
| Bedaquiline (BDQ) | 5388906 wikipedia.orgescholarship.orgresearchgate.netguidetopharmacology.orgnih.gov |
| PBTZ169 | 57331386 invivochem.comuni.lunih.govresearchgate.netcuni.cz |
Molecular Basis of Drug-drug Interactions Involving Rifampicin
Induction of Cytochrome P450 (CYP) Enzymes
Rifampicin is a well-established inducer of the cytochrome P450 (CYP) enzyme system, a superfamily of enzymes crucial for the metabolism of a vast array of endogenous and exogenous compounds, including many drugs. nih.govmdpi.com This induction leads to increased enzyme levels and activity, thereby accelerating the metabolism of co-administered drugs that are CYP substrates. mdpi.com
Transcriptional Regulation of Key CYP Isoenzymes (e.g., CYP3A4, CYP2B6, CYP2C8, CYP2C9)
This compound significantly induces the expression of several key CYP isoenzymes, with a particularly pronounced effect on CYP3A4. nih.govnih.govplos.org CYP3A4 is the most abundant CYP enzyme in the human liver and intestine and is involved in the metabolism of a large percentage of marketed drugs. mdpi.comnih.gov Studies in primary human hepatocytes have shown that this compound can increase CYP3A4 mRNA levels significantly. nih.govnih.gov this compound also induces other CYP isoforms, including CYP2B6, CYP2C8, and CYP2C9, although typically to a lesser extent than CYP3A4. nih.govmdpi.comnih.gov The induction of these enzymes by this compound occurs primarily at the transcriptional level, leading to increased synthesis of the enzyme proteins. nih.govpsu.edu
Research findings illustrate the magnitude of this compound's inductive effects on specific CYP enzymes:
In primary human hepatocytes, 20 µM this compound increased CYP3A4 mRNA by 14-fold, while CYP2B6 mRNA increased by 2.1-fold. nih.gov
In human shed enterocytes, this compound administration increased CYP3A4 mRNA expression, as well as the protein expression of CYP2C8 and CYP2C9. nih.gov
Table 1: this compound-Induced Increase in CYP Enzyme Expression
| CYP Isoenzyme | Fold Increase (mRNA/Protein) | Tissue/Cell Type | Source |
| CYP3A4 | ~14-fold (mRNA) | Primary Human Hepatocytes | nih.gov |
| CYP2B6 | ~2.1-fold (mRNA) | Primary Human Hepatocytes | nih.gov |
| CYP3A4 | Significant Increase (mRNA, protein, activity) | Human Enterocytes | nih.gov |
| CYP2C8 | Significant Increase (protein) | Human Enterocytes | nih.gov |
| CYP2C9 | Significant Increase (protein) | Human Enterocytes | nih.gov |
Activation of Nuclear Receptors: Pregnane X Receptor (PXR) and Constitutive Androstane Receptor (CAR)
The transcriptional induction of CYP enzymes by this compound is primarily mediated through the activation of nuclear receptors, particularly the Pregnane X Receptor (PXR) and, to some extent, the Constitutive Androstane Receptor (CAR). nih.govresearchgate.netpsu.edu PXR is a ligand-activated transcription factor that plays a crucial role in the regulation of genes involved in xenobiotic metabolism and transport. nih.govpsu.edu this compound is a potent agonist of human PXR. mdpi.com
Upon binding to this compound in the cytoplasm, PXR undergoes a conformational change and translocates to the nucleus. nih.govresearchgate.net In the nucleus, activated PXR forms a heterodimer with the Retinoid X Receptor (RXR). nih.govresearchgate.net This PXR/RXR heterodimer then binds to specific DNA sequences, known as xenobiotic response elements (XREs) or pregnane X receptor response elements (PXREs), located in the promoter regions of target genes, including those encoding CYP3A4, CYP2B6, CYP2C8, and CYP2C9. nih.govpsu.edu Binding of the PXR/RXR heterodimer to these response elements enhances the transcription of the downstream genes, leading to increased mRNA and protein levels of the respective CYP enzymes. nih.govpsu.edu
CAR is another nuclear receptor involved in regulating drug-metabolizing enzymes, and it can also be activated by certain xenobiotics. psu.edu While this compound is a weaker activator of CAR compared to PXR, CAR can also contribute to the induction of CYP enzymes, including CYP3A4 and CYP2B6. nih.govpsu.edu There is evidence of cross-talk between PXR and CAR in the regulation of CYP3A4 expression. psu.edu this compound-activated PXR has also been shown to interact with other nuclear receptors and coactivators, such as Hepatocyte Nuclear Factor 4 alpha (HNF4α), Steroid Receptor Coactivator-1 (SRC-1), and Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 alpha (PGC-1α), to enhance CYP3A4 gene transcription. nih.govnih.gov
Modulation of Drug Transporters
In addition to its effects on drug-metabolizing enzymes, this compound also modulates the activity and expression of various drug transporters, which are membrane proteins responsible for the uptake and efflux of drugs in various tissues, including the intestine, liver, and kidney. nih.govresearchgate.net
Impact on P-glycoprotein (P-gp) Activity and Expression
This compound is a known inducer of P-glycoprotein (P-gp), an important efflux transporter encoded by the ABCB1 (MDR1) gene. nih.govpsu.eduplos.orgresearchgate.net P-gp is located in the apical membranes of various epithelial cells, including those in the intestine, liver (canalicular membrane of hepatocytes), and blood-brain barrier, and plays a significant role in limiting the absorption and promoting the excretion of many drugs. psu.eduplos.orgresearchgate.net
The induction of P-gp by this compound occurs at the transcriptional level, mediated primarily through the activation of PXR. plos.orgacs.org PXR can bind to response elements in the ABCB1 gene promoter, leading to increased ABCB1 mRNA and subsequent P-gp protein expression. nih.govacs.org Increased P-gp expression and activity due to this compound can lead to decreased intestinal absorption and increased biliary or renal excretion of co-administered drugs that are P-gp substrates, resulting in reduced systemic exposure. nih.govresearchgate.net
Studies have demonstrated the induction of P-gp by this compound in various models:
In human duodenal biopsies, this compound treatment significantly increased P-gp content. nih.govresearchgate.net
Studies in healthy volunteers showed that this compound treatment resulted in a significant increase in duodenal P-glycoprotein content and MDR1 mRNA levels. researchgate.net
In human colon carcinoma cell lines overexpressing MDR1, this compound induction of CYP3A mRNA and protein was decreased, suggesting an influence of P-gp on intracellular this compound concentrations and subsequent CYP3A induction. pnas.org
Table 2: this compound-Induced Increase in P-glycoprotein Expression
| Transporter | Fold Increase (Protein/mRNA) | Tissue/Cell Type | Source |
| P-gp | ~3.5-fold (protein) | Human Duodenal Biopsies | nih.gov |
| P-gp | 4.2-fold (protein, Western blot), 3.5-fold (protein, immunohistochemistry) | Human Duodenal Biopsies | researchgate.net |
| P-gp | Increased (mRNA) | Human Duodenal Biopsies | researchgate.net |
| P-gp | Significant Increase (expression and activity) | RBE4 cell line (rat blood-brain barrier model) | plos.org |
While primarily an inducer, some studies suggest that this compound might also act as a substrate or even an inhibitor of P-gp under certain conditions or at specific concentrations, adding complexity to its interaction profile with P-gp substrates. plos.orgnih.gov
Effects on Glucuronosyltransferases (UGTs)
This compound also affects the activity of UDP-glucuronosyltransferases (UGTs), another important family of drug-metabolizing enzymes involved in phase II metabolism. nih.govmdpi.com UGTs catalyze the conjugation of glucuronic acid to various substrates, increasing their water solubility and facilitating their excretion. mdpi.com
Similar to CYP enzymes and P-gp, the induction of UGTs by this compound is mediated, at least in part, through the activation of nuclear receptors like PXR and CAR. nih.gov Activation of PXR and CAR can lead to increased transcription of certain UGT genes, such as UGT1A. nih.gov This induction can result in increased glucuronidation and thus enhanced clearance of drugs that are UGT substrates. nih.govmdpi.com
Table 3: this compound's Effects on UGTs
| Enzyme Family | Effect | Mechanism Involved | Source |
| UGTs | Induction | PXR/CAR activation | nih.gov |
| UGT1A | Induction | PXR/CAR mediated | nih.gov |
| UGT1A4 | Inhibition (in vitro) | Not fully elucidated | nih.gov |
The complex interplay between this compound and these drug-metabolizing enzymes and transporters underscores the importance of considering potential drug-drug interactions when co-administering this compound with other medications. nih.govresearchgate.net
Pharmacokinetic and Pharmacodynamic Research Approaches for Rifampicin Theoretical/modeling
Population Pharmacokinetic (PopPK) Modeling
Population pharmacokinetic modeling is a powerful tool used to quantify and understand the sources of variability in drug concentrations among individuals within a target population. nih.govdovepress.comdovepress.comasm.org This approach involves analyzing drug concentration data collected from a group of individuals simultaneously, allowing for the estimation of typical population pharmacokinetic parameters and their inter-individual variability. nih.govdovepress.comdovepress.comasm.org
Most rifampicin PopPK models have employed a one-compartment linear elimination model. nih.govdovepress.comtandfonline.comdovepress.com To better describe the absorption phase of this compound, which can be complex, models often incorporate transit compartments or lagged absorption. nih.govdovepress.comtandfonline.comdovepress.comresearchgate.net A one-compartment model with first-order absorption and a lag time has been used to describe this compound plasma concentrations. asm.org Some studies have also utilized a two-compartment model to describe this compound pharmacokinetics. dovepress.comoup.com For instance, a two-compartment disposition model with first-order transit oral absorption and elimination via saturable hepatic extraction has been identified as providing a good description of this compound pharmacokinetics in plasma and cerebrospinal fluid in adults with tuberculosis meningitis. oup.com A model incorporating saturable pharmacokinetics and autoinduction for high this compound doses also utilized a one-compartment disposition model with a transit compartment absorption model. nih.govoup.com
PopPK modeling allows for the identification and quantification of covariates that contribute to the observed variability in this compound pharmacokinetics. nih.govdovepress.comtandfonline.comdovepress.comresearchgate.net Body weight (BW) is a frequently identified covariate influencing this compound PK parameters, including clearance and volume of distribution. nih.govdovepress.comtandfonline.comdovepress.comresearchgate.net An allometric growth model based on fat-free mass (FFM) has also been explored and might improve model fit. nih.govdovepress.comtandfonline.comresearchgate.net Postmenstrual age (PMA) has been found to significantly impact elimination in pediatric patients. nih.govdovepress.comtandfonline.comdovepress.com Other covariates that have been identified as significant in affecting this compound PK parameters include pharmaceutical formulation, gender, pregnancy status, diabetes, and nutritional supplementation. researchgate.net HIV status has also been investigated, with some studies indicating it can affect serum concentrations. nih.govtandfonline.com
Here is a table summarizing some identified covariates and their potential impact:
| Covariate | Effect on PK Parameters (Examples) | Source(s) |
| Body Weight (BW) | Influences clearance and volume of distribution. | nih.govdovepress.comtandfonline.comdovepress.comresearchgate.net |
| Fat-Free Mass (FFM) | Might improve model fit, potentially relevant for dosing optimization. | nih.govdovepress.comtandfonline.comresearchgate.net |
| Postmenstrual Age (PMA) | Significantly impacts elimination in pediatrics. | nih.govdovepress.comtandfonline.comdovepress.com |
| Gender | Identified as a significant covariate. | researchgate.net |
| Pregnancy Status | Identified as a significant covariate. | researchgate.net |
| Diabetes | Identified as a significant covariate. | researchgate.net |
| Nutritional Supplementation | Identified as a significant covariate. | researchgate.net |
| HIV Status | Can lead to reduced serum concentrations. | nih.govtandfonline.com |
Modeling and simulation techniques are employed to analyze the variability in this compound exposure within a population and to predict exposure under different scenarios. nih.govdovepress.comdovepress.comasm.org Significant variations in exposure predictions have been observed among different PopPK models, highlighting challenges in achieving therapeutic targets with standard treatment approaches. nih.govdovepress.comtandfonline.comdovepress.com Simulations have shown that the exposure of patients receiving the same dose can vary considerably, even with weight-based dosing. frontiersin.org Monte Carlo simulations are often used to estimate optimal dosing strategies and predict exposure profiles in virtual patient populations. dovepress.comasm.orgnih.govresearcher.life These simulations can quantify inter-individual variability (IIV) and inter-occasion variability (IOV) in exposure parameters like AUC. frontiersin.org For example, simulations have shown that IOV in AUC for this compound can be as high as the IIV. frontiersin.org
Identification and Quantification of Covariates Affecting PK Parameters (e.g., body weight, post-natal age)
Pharmacodynamic Relationship Research
Pharmacodynamic relationship research focuses on understanding the link between this compound exposure and its pharmacological effect, particularly its antibacterial activity against Mycobacterium tuberculosis. asm.orgmdpi.comtandfonline.com This involves developing models that describe how drug concentrations relate to measures of efficacy. asm.orgmdpi.comtandfonline.com
Exposure-response modeling is conducted in preclinical and research models, such as in vitro systems and animal models, to characterize the relationship between this compound exposure and its effect on bacterial growth and killing. mdpi.comnih.govnih.govoup.comresearchgate.net The ratio of the area under the concentration-time curve from 0 to 24 hours (AUC₀₋₂₄) to the minimum inhibitory concentration (MIC) is considered a key pharmacokinetic-pharmacodynamic parameter for predicting efficacy. asm.orgtandfonline.commdpi.com Preclinical studies, including those using hollow-fiber infection models and murine models, have demonstrated that the AUC/MIC ratio is the best predictive PD parameter for this compound efficacy. asm.org The Multistate Tuberculosis Pharmacometric (MTP) model is a semi-mechanistic model used to describe and identify the exposure-response profile of antituberculosis drugs, including this compound, against different bacterial subpopulations in vitro and in vivo. mdpi.comnih.govnih.govoup.comresearchgate.net This model has been used to predict this compound biomarker response in hollow-fiber infection models, murine studies, and early clinical trials. mdpi.comnih.govnih.govresearchgate.net
Predictive modeling aims to translate the understanding of PK/PD relationships into strategies for achieving optimal this compound exposure in patients. asm.orgmdpi.comnih.gov Model-informed approaches are used to predict the exposures needed for desired effects and to guide dose selection. mdpi.comnih.gov For instance, model-based meta-analysis has been used to evaluate different this compound doses and predict the probability of attaining target exposures associated with improved outcomes. nih.gov Predictive models can also be used to simulate concentration-time profiles and assess whether standard or alternative dosing strategies are likely to achieve optimal exposure in various patient populations. nih.gov The goal is to use these models to support individualized dosing and improve treatment outcomes by ensuring adequate drug exposure. nih.govdovepress.comdovepress.comasm.org Predictive modeling, including approaches utilizing machine learning, is being explored to determine individualized initial doses and improve the probability of target attainment. researchgate.net
Advanced Analytical Methodologies for Rifampicin Research
Chromatographic Techniques
Chromatographic techniques play a crucial role in separating Rifampicin from complex matrices and its related substances or impurities, enabling accurate quantification and characterization.
High-Performance Liquid Chromatography (HPLC) with UV Detection
High-Performance Liquid Chromatography coupled with Ultraviolet (UV) detection is a widely utilized method for the analysis of this compound in various samples, including pharmaceutical formulations, bulk drug, and biological fluids like plasma. This technique offers advantages in terms of sensitivity, accuracy, and reproducibility.
Numerous research studies have detailed HPLC-UV methods for this compound analysis. A validated RP-HPLC method with UV detection at 219 nm was developed for routine analysis of this compound in pharmaceutical formulations, demonstrating linearity over a concentration range of 100-600 µg/ml with a correlation coefficient of 0.9957. The method showed good precision with a relative standard deviation (RSD) of 0.20% for intraday measurements. researchgate.net Another HPLC method for this compound analysis in plasma utilized a UV-Vis detector at 377 nm with a mobile phase of acetonitrile-phosphate buffer pH 6.8 (45:55) at a flow rate of 1.5 ml/min. This method was validated according to ICH and EMA guidelines, showing linearity from 0.05 to 10.26 µg/ml (R² = 0.9984), with precision (%RSD) between 1.40% and 13.04% and accuracy (%Recovery) between 86.24% and 102.13%. innovareacademics.in
HPLC-UV methods have also been developed for the simultaneous determination of this compound alongside other drugs. For instance, a method for simultaneous analysis of this compound and clindamycin phosphate in skin layers used a C18 column with gradient elution and UV-Vis detection at 238 nm for this compound. This method was linear from 0.5 to 20.0 µg/mL (r² > 0.999) with recovery rates exceeding 85%. nih.gov Another study for simultaneous determination of this compound, Isoniazid, and Pyrazinamide in human plasma employed reversed-phase HPLC with UV detection at 344.0 nm, showing linearity for this compound from 2.5 to 35.0 μg/mL with good accuracy and precision. frontiersin.org
Several studies highlight specific chromatographic conditions for this compound analysis by HPLC-UV:
| Column Type | Mobile Phase | Detection Wavelength (nm) | Flow Rate (mL/min) | Matrix | Linearity Range (µg/ml) | Ref. |
| C18 | Acetonitrile-phosphate buffer pH 6.8 (45:55) | 377 | 1.5 | Human Plasma | 0.05 - 10.26 | innovareacademics.in |
| Primesep 100 | MeCN – 70%, Ammonium Formate pH 3.0-20 mM Buffer | 270 | 0.2 | - | - | sielc.com |
| Symmetry Syncronis C18 | Phosphate buffer (A) and acetonitrile (B) (50:50) | 236 | 1.0 | Bulk and Pharmaceutical Formulations | 25 - 125 | rjptonline.org |
| Chromolith RP8 | 0.05 M acetate buffer pH 5.7-acetonitrile (35:65) | 335 | - | Human Plasma | 0.05 - (mg/L) | nih.gov |
| Zorbax Rx C8 | 0.05 M potassium dihydrogen phosphate-acetonitrile (55:45) | 340 | - | Human Plasma | 0.05 - 35 (µg/ml) | researchgate.net |
| C18 (150 mm x 4.6 mm, 5 µm) | 0.01 M phosphoric acid and methanol (gradient) | 238 | 1.0 | Skin layers (with Clindamycin phosphate) | 0.5 - 20.0 | nih.gov |
These studies demonstrate the versatility of HPLC-UV for quantifying this compound in diverse sample types, supporting research in areas such as bioequivalence and drug distribution.
Liquid Chromatography-Mass Spectrometry (LC-MS/MS)
Liquid Chromatography-Mass Spectrometry (LC-MS/MS) offers enhanced sensitivity and specificity compared to HPLC-UV, making it particularly valuable for the analysis of this compound in complex biological matrices and for the detection of impurities and metabolites. LC-MS/MS provides molecular specificity, allowing for rapid differentiation between drugs and their metabolites. researchgate.netijper.org
A highly sensitive and rapid LC-MS/MS method was developed and validated for the quantification of this compound in human plasma and cerebrospinal fluid (CSF). researchgate.netnih.gov This method involved a simple protein precipitation step for sample preparation and detection was performed by electrospray positive ionization mass spectrometry in multiple-reaction monitoring (MRM) mode. researchgate.netnih.gov The method was linear in the concentration range of 25–6400 ng/mL with intra- and inter-day precision below 7% and 8%, respectively. nih.gov Another fast and simple LC-MS/MS method for this compound determination in human blood plasma utilized protein precipitation and a C18 column, with a total run time of 4 minutes. Detection was performed by monitoring the m/z of the parent ion at 823.691 and a product ion at 791.757. ijper.org
LC-MS/MS is also applied for the determination of impurities in this compound. A method was developed for the determination of 1-Methyl-4-Nitrosopiperazine (MNP), a genotoxic impurity, in this compound products using LC-MS/MS. This method required extraction with neutralization and was validated according to regulatory guidelines, evaluating parameters such as specificity, accuracy, precision, LOD, and LOQ. mdpi.com High-resolution and high-mass accuracy LC-ESI-HRMS methods are used to separate and detect nitrosamine impurities like MNP in this compound, achieving high sensitivity by monitoring the accurate m/z of fragment ions. fda.gov
The application of LC-MS/MS for this compound analysis in biological matrices is crucial for pharmacokinetic studies and therapeutic drug monitoring, offering the required sensitivity and selectivity.
High-Performance Thin-Layer Chromatography (HPTLC)
High-Performance Thin-Layer Chromatography (HPTLC) is a planar chromatographic technique that can be used for the analysis of this compound, often in combination with other drugs. HPTLC offers advantages in terms of simplicity and the ability to analyze multiple samples simultaneously on a single plate.
HPTLC methods have been reported for the simultaneous determination of this compound and Isoniazid in rat plasma and pharmaceutical formulations. One method used silica gel 60 F254 plates with a mobile phase of ethyl acetate–methanol–acetone–acetic acid (5 + 2 + 2 + 1, v/v). Quantitative analysis was performed by UV-visible densitometric scanning at wavelengths specific to each compound (details on specific wavelengths for this compound in this method were not explicitly provided in the snippet, but other sources indicate UV detection for HPTLC of this compound). akjournals.com Another HPTLC method for simultaneous estimation of this compound and Isoniazid in rat plasma used silica gel 60 F254 plates and a mixture of chloroform:methanol as the mobile phase, with densitometer absorbance mode detection at 475 nm for this compound and 280 nm for Isoniazid. This method had a linearity range of 10 to 80 µg/ml for both drugs and a detection limit of 0.3 µ g/spot for this compound. ijpsonline.com
HPTLC methods, while perhaps less sensitive than LC-MS/MS, provide a valuable tool for the analysis of this compound, particularly in combination formulations and for screening purposes.
Hyphenated Techniques (e.g., GC-MS, LC-NMR, LC-IR)
Hyphenated techniques combine separation methods with spectroscopic detection to enhance the analytical capabilities for complex samples. These techniques are particularly useful for impurity profiling and structural elucidation. Common examples include GC-MS, LC-MS, LC-NMR, and LC-IR. monash.educhromatographytoday.comajrconline.orgsaspublishers.comnih.gov
While LC-MS/MS has been discussed as a powerful hyphenated technique for this compound quantification and impurity detection researchgate.netijper.orgnih.govmdpi.comfda.gov, other hyphenated techniques like LC-NMR and LC-IR are primarily mentioned in the broader context of their utility in pharmaceutical analysis and impurity characterization rather than providing specific detailed research findings on their application solely to this compound.
LC-NMR, for instance, is a powerful complement to LC-UV-MS for screening and provides structural information. nih.gov LC-IR combines liquid chromatography with infrared spectrometry, which is useful for identifying organic compounds based on their characteristic absorption bands. ajrconline.orgnih.gov GC-MS is another hyphenated technique, but it is generally more suitable for volatile and thermally stable molecules, which may limit its direct application to this compound, a relatively large and less volatile molecule. monash.edu
Although specific detailed studies focusing solely on the application of GC-MS, LC-NMR, or LC-IR for the analysis of this compound itself were not prominently found in the provided search results, these techniques are valuable tools in pharmaceutical research for the comprehensive characterization of drug substances and impurities, which would indirectly support this compound research by enabling the analysis of related compounds and degradation products.
Spectroscopic Methods
Spectroscopic methods analyze the interaction of light or other radiation with a substance to gain information about its structure and concentration.
UV-Vis Spectrometry
UV-Vis Spectrometry is a simple and cost-effective technique widely used for the quantitative analysis of this compound in various matrices, including bulk drug, pharmaceutical formulations, and biological samples. This method relies on the absorption of UV or visible light by the this compound molecule at specific wavelengths.
Numerous studies have reported the use of UV-Vis spectrophotometry for this compound quantification. A simple UV spectrophotometric method for determining this compound in bulk and capsules showed a detection wavelength at 337 nm and followed Beer's law over the concentration range of 5-13 µg/ml. The method demonstrated good accuracy with percentage recoveries between 98-100%. ajpamc.com Another visible spectrophotometric method for this compound estimation in bulk and pharmaceutical dosage forms showed maximum absorbance at 475 nm, with linearity in the concentration range of 10-30 µg/ml and a correlation of 0.999. pharmacyjournal.net
UV-Vis spectrophotometry has also been validated for quantifying this compound in biological matrices and different media. A validated UV-Vis spectrophotometric colorimetric method for this compound quantification in phosphate-buffered saline (PBS), plasma, and brain tissue demonstrated excellent linearity (r² = 0.999) and detection limits ranging from approximately 0.25–0.49 μg/mL. tandfonline.comnih.gov The method showed high accuracy and precision within acceptable ranges. tandfonline.comnih.gov
The UV-Vis spectrum of this compound in water typically shows two absorption bands at approximately 333 nm and 472 nm. researchgate.net The specific wavelength of maximum absorbance can vary slightly depending on the solvent or buffer used. For instance, in phosphate buffer pH 7.0, a maximum absorption is observed at 470 nm. In methanol and phosphate buffer pH 7.4, a peak wavelength of 474 nm is reported. rjptonline.org
UV-Vis spectrophotometry, despite being less selective than chromatographic methods, remains a valuable tool for routine analysis and quantification of this compound due to its simplicity, speed, and cost-effectiveness.
Infrared (IR) Spectroscopy
Infrared (IR) spectroscopy is a valuable tool for the characterization of this compound, particularly in identifying functional groups and assessing its solid-state form, including polymorphism. IR spectroscopy can detect major absorption bands characteristic of this compound. For instance, studies have shown distinct absorption bands for the OH group of the chain loop, acetyl group, and furanone group, which can differentiate between polymorphic forms of this compound. iajps.com The IR spectrum of this compound Form I, for example, shows characteristic bands at approximately 3400 cm⁻¹, 1722 cm⁻¹, and 1643 cm⁻¹. iajps.com Form II exhibits different bands at 3356 cm⁻¹, 1732 cm⁻¹, and 1714 cm⁻¹. iajps.com FTIR has also been used to investigate the interaction between this compound and other substances, such as polymers in nanosuspensions or materials in drug delivery systems, by observing shifts or changes in characteristic peaks, confirming the presence of the drug and indicating potential interactions like hydrogen bonding or electrostatic interactions without forming new compounds. nih.govdovepress.comrsc.orgbrieflands.com
Nuclear Magnetic Resonance (NMR) Spectroscopy (1D and 2D)
Nuclear Magnetic Resonance (NMR) spectroscopy, including both one-dimensional (1D) and two-dimensional (2D) techniques, provides detailed structural information about this compound. These methods are essential for assigning ¹H and ¹³C NMR signals and elucidating the complete molecular structure. 2D NMR techniques such as ¹H-¹H correlated spectroscopy (COSY) and ¹H-¹³C heteronuclear multiple-bond correlation (HMBC) are particularly useful for establishing connectivity and spatial relationships within the complex this compound molecule. researchgate.netresearchgate.net NMR analysis has been used to confirm the structure of this compound and its derivatives and to study chemical transformations, such as the linearization of rifamycin antibiotics catalyzed by certain enzymes. frontiersin.orgfrontiersin.org Intensive 1D and 2D NMR analyses have unambiguously demonstrated such enzymatic activities and helped elucidate the structure of reaction products. frontiersin.orgfrontiersin.org Beyond structural determination, NMR metabolomics, combining 1D ¹H NMR with multivariate analysis, has been explored to characterize metabolic profiles in biological systems treated with antibiotics like this compound, aiding in understanding their effects at a molecular level. nih.gov
Luminescence-Based Methods (e.g., Chemiluminescence, Fluorescence)
Luminescence-based methods, including chemiluminescence and fluorescence, offer sensitive approaches for the detection and analysis of this compound. These methods often rely on the inherent luminescence properties of this compound or its interaction with luminescent probes. Fluorescence quenching, for instance, has been utilized for the detection of residual this compound. A method involving the fluorescence quenching of gold nanoclusters stabilized by bovine serum albumin (BSA-Au NCs) has been developed for detecting this compound in urine samples. nih.gov The fluorescence intensity of the BSA-Au NCs decreases proportionally with increasing this compound concentration, showing a nearly linear relationship over a specific range. nih.gov This allows for sensitive detection, with reported detection limits as low as 70 ng/mL. nih.gov Luminescence methods are summarized as part of various optical and luminescence techniques used for rifamycin antibiotics analysis. mdpi.comresearchgate.netnih.govsemanticscholar.orgsemanticscholar.orgdntb.gov.ua
Electrochemical Methods
Electrochemical methods are widely applied for the analysis of this compound, offering advantages in terms of sensitivity, simplicity, and cost-effectiveness. These techniques involve measuring the electrical properties of a solution containing this compound as a function of applied potential or current. mdpi.comresearchgate.netnih.govsemanticscholar.orgsemanticscholar.org
Voltammetry, Amperometry, Potentiometry, Photoelectrochemistry
Various electrochemical techniques, including voltammetry, amperometry, potentiometry, and photoelectrochemistry, have been employed for the determination of this compound. Voltammetry, particularly cyclic voltammetry, is frequently used to characterize the electrochemical behavior of this compound, revealing its oxidation and reduction peaks. mdpi.comresearchgate.net Studies using cyclic voltammetry with modified electrodes, such as glassy carbon electrodes modified with carbon nanotubes, have been conducted to analyze this compound in different media, including blood. researchgate.net These studies investigate the current peaks at different concentrations, pH levels, and scan rates to understand the electrochemical properties of this compound. researchgate.net Amperometric, potentiometric, and photoelectrochemical methods, often coupled with sensors, are also utilized for the analysis of this compound and other rifamycin antibiotics in various matrices, including pharmaceutical formulations, biological fluids, and environmental samples. mdpi.comresearchgate.netnih.govsemanticscholar.orgsemanticscholar.org
Method Validation and Application in Research Settings
The validation of analytical methods for this compound is a critical step to ensure their reliability, accuracy, and suitability for intended research applications. Validation studies are performed according to established guidelines, such as those from the ICH. ipinnovative.comcore.ac.ukajpamc.com
Specificity, Sensitivity, Linearity, Accuracy, Precision, Robustness Studies
Method validation for this compound analysis involves evaluating several key performance characteristics.
Specificity: This confirms that the method can accurately measure this compound in the presence of other components that may be present in the sample matrix, such as excipients in pharmaceutical formulations or endogenous substances in biological fluids. ipinnovative.comcore.ac.ukajpamc.comresearchgate.net
Sensitivity: This is determined by the Limit of Detection (LOD) and Limit of Quantification (LOQ), which represent the lowest concentrations of this compound that can be reliably detected and quantified, respectively. ipinnovative.comajpamc.comresearchgate.netnih.gov
Linearity: This assesses the proportional relationship between the this compound concentration and the analytical response over a defined range. A high correlation coefficient (r²) indicates good linearity. ipinnovative.comajpamc.comresearchgate.netnih.gov
Accuracy: This measures how close the experimental results are to the true concentration of this compound. It is often evaluated through recovery studies by analyzing samples with known concentrations. ipinnovative.comcore.ac.ukajpamc.comresearchgate.netnih.gov
Precision: This indicates the reproducibility of the method under the same conditions (repeatability) and under different conditions, such as different days or analysts (intermediate precision/ruggedness). It is typically expressed as relative standard deviation (% RSD). ipinnovative.comcore.ac.ukajpamc.comresearchgate.netnih.gov
Robustness: This evaluates the method's ability to remain unaffected by small, deliberate variations in method parameters. ipinnovative.comajpamc.comresearchgate.net
Validated methods are then applied in various research settings, including studies on drug delivery systems, drug interactions, and the analysis of this compound in complex matrices. nih.govipinnovative.comresearchgate.netnih.gov For example, validated HPLC methods have been used for the simultaneous determination of this compound with other compounds or in pharmaceutical formulations. ipinnovative.comcore.ac.ukresearchgate.net LC-MS/MS methods have been validated for the determination of this compound and its metabolites in human plasma to support clinical trials. nih.gov
Here is an example of validation data for a specific RP-HPLC method for this compound:
| Validation Parameter | Result | Acceptance Criteria (ICH Q2R1) | Source |
|---|---|---|---|
| System Precision (% RSD) | 0.9% (for 6 sample solutions) | ≤ 2.0% | ipinnovative.com |
| Specificity | No interference from blank, placebo, or known impurities | Peak purity passed | ipinnovative.com |
| Linearity (Correlation Coefficient, r) | 1.000 | ≥ 0.999 | ipinnovative.com |
| Accuracy (% Recovery) | 101.1% (80%), 100.4% (100%), 100.5% (120%) | 98-102% | ipinnovative.com |
| Method Precision (% RSD) | Satisfactory | ≤ 2.0% | ipinnovative.comcore.ac.uk |
| Intermediate Precision (% RSD) | Satisfactory | ≤ 2.0% | core.ac.uk |
| LOQ | 0.18 ppm | - | ipinnovative.com |
Another example shows validation parameters for an RP-HPLC method for simultaneous determination of this compound and a flavonoid glycoside:
| Validation Parameter | This compound Result | Flavonoid Glycoside Result | Source |
|---|---|---|---|
| Linearity (r²) | > 0.999 | > 0.999 | researchgate.net |
| Precision (% RSD) | 1.08-2.77% | 1.14-2.98% | researchgate.net |
| LOQ | 0.10 µg/mL | 0.05 µg/mL | researchgate.net |
| Robustness | Method found robust | Method found robust | researchgate.net |
| Specificity | Method found specific | Method found specific | researchgate.net |
Application in In Vitro and In Vivo (Animal) Model Studies
This compound, a key antibiotic, has been extensively studied using both in vitro and in vivo animal models to understand its mechanism of action, efficacy against various pathogens, and the development of resistance. These studies are crucial for evaluating its potential therapeutic applications and optimizing treatment strategies.
In vitro studies have provided fundamental insights into how this compound inhibits bacterial growth. This compound exerts its effect by binding to the beta-subunit of bacterial DNA-dependent RNA polymerase (RNAP), thereby preventing the initiation of RNA synthesis wikipedia.orgdrugbank.compatsnap.com. This binding is highly selective for bacterial RNAP and does not affect the mammalian enzyme drugbank.compatsnap.com. Studies using Escherichia coli have shown that this compound inhibits both RNA and protein synthesis, with the rates decreasing exponentially after an initial lag phase nih.gov. Higher external concentrations of this compound were needed to achieve comparable inhibition in normal E. coli cells compared to those treated with ethylenediaminetetraacetic acid (EDTA), suggesting differences in cell permeability nih.gov. The kinetics of inhibition were found to be concentration-dependent below certain this compound concentrations nih.gov.
This compound's activity against bacterial biofilms has also been investigated in vitro. Studies have shown promising antibiofilm effects against Staphylococcus aureus, particularly when used in combination with other antibiotics like minocycline or tigecycline frontiersin.org. This antibiofilm activity has been consistently demonstrated against Staphylococcus species, including MRSA and S. epidermidis, often as part of combination regimens frontiersin.org. While some studies report good efficacy against in vitro biofilms under favorable conditions, this does not always translate to efficacy in animal models nih.gov.
Animal models, predominantly mice, rats, and rabbits, have been instrumental in evaluating the in vivo efficacy of this compound against various infections, including tuberculosis (TB) and staphylococcal infections nih.govfrontiersin.orgjci.orgd-nb.infoasm.orgvivexia.fracs.org. These models allow for the assessment of this compound's activity within a complex biological system, considering factors like pharmacokinetics, tissue distribution, and the host immune response.
Murine models of tuberculosis have been widely used to study this compound's effectiveness. High-dose this compound regimens have shown improved bactericidal activity and reduced relapse rates in murine TB models compared to standard doses frontiersin.orgjci.org. Studies have demonstrated a dose-dependent eradication of Mycobacterium tuberculosis, including persistent bacteria, in these models frontiersin.org. Increasing the dose of this compound also significantly reduced the risk of antibiotic resistance emergence in a murine infection model frontiersin.org. In the Cornell mouse model, a high-dose this compound regimen resulted in faster visceral clearance of M. tuberculosis frontiersin.org.
| Treatment Regimen (Murine TB Model) | Time to Organ Sterility (Weeks) | Relapse Rate (%) |
| Standard-dose this compound | 14 | 87.5 frontiersin.org |
| High-dose this compound (50 mg/kg) | 8 | 0 frontiersin.org |
Animal models have also been crucial for studying this compound's efficacy against staphylococcal infections, particularly those involving biofilms and implanted medical devices nih.govvivexia.fr. The tissue-cage infection model in guinea pigs has been used to test the efficacy of antimicrobial agents against foreign body-associated infections nih.gov. In this model, combination therapy including this compound has shown high efficacy against Staphylococcus species nih.gov. Studies in rat models of methicillin-resistant Staphylococcus epidermidis (MRSE) osteomyelitis have shown that this compound monotherapy rapidly reduced bacterial load in bone tissue, and combination with ceftaroline further shortened the duration of positive cultures vivexia.fr.
| Treatment (Rat MRSE Osteomyelitis Model) | Mean Tibial Bacterial Load (log10 CFU/g) at Day 3 | Mean Tibial Bacterial Load (log10 CFU/g) at Day 7 |
| Saline Control | - | 4.4 vivexia.fr |
| Rifampin Alone | 1.89 ± 1.0 vivexia.fr | Limit of detection vivexia.fr |
| Ceftaroline Alone | 3.8 ± 0.6 vivexia.fr | 3.6 vivexia.fr |
| Ceftaroline + Rifampin | 1.68 ± 1.0 vivexia.fr | Limit of detection vivexia.fr |
Studies in rabbits with experimentally induced TB meningitis have also shown that high-dose this compound regimens lead to significantly higher bactericidal activity in brain tissues compared to standard doses jci.org.
Research involving inhaled formulations of this compound in animal models like rats and guinea pigs suggests that this delivery method can achieve higher drug concentrations in the lungs and alveolar macrophages compared to oral administration d-nb.infonih.gov. These preclinical studies support the potential of inhaled this compound for treating pulmonary TB d-nb.infonih.gov.
In vivo studies have also explored the use of this compound-loaded nanoparticles to enhance therapeutic efficacy against infections like Brucella canis. In a murine model, this compound-loaded PLGA nanoparticles demonstrated superior effectiveness in reducing bacterial load in the spleen compared to the free drug acs.org.
These in vitro and in vivo animal model studies collectively contribute significantly to the understanding of this compound's antimicrobial properties, informing the development of more effective treatment strategies and formulations.
Q & A
Q. What are the standard methodologies for detecting rifampicin resistance in Mycobacterium tuberculosis?
The Xpert® MTB/RIF assay is a widely validated molecular test for simultaneous TB diagnosis and this compound resistance detection. It employs real-time PCR to amplify the rpoB gene and identify mutations linked to resistance. Sensitivity ranges from 88% (as an initial test) to 67% (as an add-on after negative microscopy), with specificity >98% . Culture-based drug susceptibility testing (DST) remains the gold standard but requires weeks for results. For epidemiological studies, resistance trends can be analyzed using χ2 trend tests and spatial mapping tools like GeoDa to visualize regional patterns .
Q. How is this compound content quantified in pharmaceutical formulations?
Near-infrared (NIR) diffuse reflectance spectroscopy is a rapid, non-destructive method. Researchers develop calibration models by correlating spectral data with HPLC-validated this compound concentrations. Key steps include spectral preprocessing (e.g., Savitzky-Golay smoothing), partial least squares regression for model construction, and validation using root mean squared error (RMSE) metrics .
What frameworks guide the formulation of research questions in this compound studies?
The PICOT framework (Population, Intervention, Comparison, Outcome, Time) is effective. For example:
- Population: Adults with this compound-resistant TB in high-burden regions.
- Intervention: Novel this compound-nanoparticle delivery.
- Comparison: Standard oral this compound.
- Outcome: Reduction in treatment duration.
- Time: 6-month follow-up. This structure ensures alignment with gaps in pharmacokinetics or resistance mechanisms .
Q. How are in vitro CYP induction studies standardized using this compound?
this compound serves as a positive control in CYP3A4 induction assays. Data normalization involves expressing test compound fold-induction as a percentage of this compound’s maximal response (e.g., Emax). Curve-fitting models (e.g., sigmoidal dose-response) calculate EC50 values. Compliance with this compound pretreatment protocols is critical to avoid skewed results due to auto-induction variability .
Advanced Research Questions
Q. What experimental designs optimize pH-responsive this compound delivery systems?
A 2⁴ factorial design can evaluate variables like adsorbent mass, pH, and drug concentration. For example, palygorskite clay nanocarriers achieved 33.62 mg/g drug loading at pH 2. Post-synthesis, kinetic models (e.g., pseudo-second-order) and isotherms (e.g., Langmuir) elucidate adsorption mechanisms. Characterization via FTIR, XRD, and SEM confirms drug-clay interactions and amorphous state stabilization .
Q. How can Box-Behnken designs improve this compound nanoparticle formulations?
Box-Behnken response surface methodology (3 factors, 3 levels) optimizes variables like lecithin-oleic acid concentration (X1), this compound dose (X2), and polysorbate 80 (X3). Responses include particle size (Y1) and entrapment efficiency (Y2). Minitab-generated polynomial equations identify significant interactions, e.g., reducing particle size by balancing surfactant and drug ratios .
Q. What bioinformatics tools analyze this compound’s transcriptional effects in bacteria?
The R package rifi processes RNA-seq or microarray time-series data from this compound-treated cultures. Steps include:
Q. How do researchers resolve contradictions in this compound’s CYP induction data?
Discrepancies arise from variability in cell lines (e.g., hepatocytes vs. transfected cells) and this compound exposure protocols. Meta-analyses should stratify data by experimental conditions. Sensitivity analyses exclude outliers (e.g., subjects with 100-fold higher this compound concentrations due to non-compliance) .
Q. What spatial-temporal methods track this compound resistance epidemiology?
GeoDa software maps resistance rates using Moran’s I for spatial autocorrelation. Temporal trends are assessed via χ2 trend tests in SPSS. For instance, Zhejiang Province (2015–2019) reported 5.9% this compound resistance, with hotspots linked to urbanization and healthcare worker exposure .
Q. How are phase 1 trials designed for inhalable this compound formulations?
Dry powder inhalers require particle size optimization (<5 µm for alveolar deposition). Phase 1 trials assess pharmacokinetics (Cmax, AUC) and safety via spirometry and bronchoalveolar lavage. Recruitment criteria exclude patients with chronic respiratory conditions. Data are analyzed using non-compartmental models in Phoenix WinNonlin .
Featured Recommendations
| Most viewed | ||
|---|---|---|
| Most popular with customers |
Disclaimer and Information on In-Vitro Research Products
Please be aware that all articles and product information presented on BenchChem are intended solely for informational purposes. The products available for purchase on BenchChem are specifically designed for in-vitro studies, which are conducted outside of living organisms. In-vitro studies, derived from the Latin term "in glass," involve experiments performed in controlled laboratory settings using cells or tissues. It is important to note that these products are not categorized as medicines or drugs, and they have not received approval from the FDA for the prevention, treatment, or cure of any medical condition, ailment, or disease. We must emphasize that any form of bodily introduction of these products into humans or animals is strictly prohibited by law. It is essential to adhere to these guidelines to ensure compliance with legal and ethical standards in research and experimentation.


