Vicriviroc
Description
Synthesis of Vicriviroc
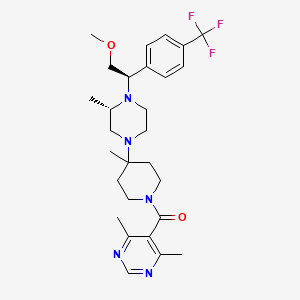
The chemical synthesis of this compound (SCH 417690) has been previously described nih.gov. This compound has the chemical formula C₂₈H₃₈F₃N₅O₂ and a molecular weight of 533.29775996 Da nih.govbocsci.comwikidata.org. Its chemical name is (4,6-dimethylpyrimidin-5-yl)-[4-[(3S)-4-[(1R)-2-methoxy-1-[4-(trifluoromethyl)phenyl]ethyl]-3-methylpiperazin-1-yl]-4-methylpiperidin-1-yl]methanone nih.govbocsci.com.
Studies have also involved the synthesis of isotopically labeled forms of this compound for research purposes, such as absorption, distribution, metabolism, and excretion (ADME) studies and CCR5 receptor binding work nih.gov. For example, low specific activity [³H]SCH 417690 was prepared for preliminary ADME evaluations, while [¹⁴C]SCH 417690 was synthesized for more definitive ADME work, including human studies nih.gov. High specific activity [³H]SCH 417690 was prepared for receptor binding studies, and [²H₄]SCH 417690 was synthesized for use as an internal standard in liquid chromatography-mass spectrometry bioanalytical methods nih.gov.
Development of Analogues for Enhanced Activity or Specificity
The development of this compound involved structural optimization based on earlier CCR5 inhibitors like SCH-C bocsci.com. This compound itself was selected from a series of piperazine core compounds based on their activity in binding and antiviral assays nih.gov. The search for isosteric replacements for certain groups, such as the n-propyl group in related compounds, led to the identification of analogues, including this compound (SCH-417690), which demonstrated excellent potency against R5-tropic HIV and favorable oral bioavailability in animal models acs.orgresearchgate.net.
Structure-activity relationship studies were crucial in the discovery and optimization of CCR5 antagonists wikipedia.orgacs.orgdntb.gov.ua. These studies aimed to identify compounds with enhanced activity, improved pharmacokinetic properties, and better selectivity for CCR5 over other receptors like muscarinic receptors and hERG wikipedia.orgnih.govacs.org. The enhanced selectivity observed with this compound compared to SCH-C was reflected in an improved side effect profile during safety pharmacology evaluations acs.org.
The development of analogues is a continuous process in medicinal chemistry to identify compounds with optimized properties. While specific detailed data tables on the synthesis and analogue development of this compound were not extensively found in the search results, the process inherently involves the synthesis and evaluation of various chemical structures to determine their biological activity, binding affinity, and pharmacokinetic profiles, guiding the selection and optimization of lead compounds like this compound wikipedia.orgnih.govacs.orgresearchgate.net.
Properties
IUPAC Name |
(4,6-dimethylpyrimidin-5-yl)-[4-[(3S)-4-[(1R)-2-methoxy-1-[4-(trifluoromethyl)phenyl]ethyl]-3-methylpiperazin-1-yl]-4-methylpiperidin-1-yl]methanone | |
|---|---|---|
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI |
InChI=1S/C28H38F3N5O2/c1-19-16-35(14-15-36(19)24(17-38-5)22-6-8-23(9-7-22)28(29,30)31)27(4)10-12-34(13-11-27)26(37)25-20(2)32-18-33-21(25)3/h6-9,18-19,24H,10-17H2,1-5H3/t19-,24-/m0/s1 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI Key |
CNPVJJQCETWNEU-CYFREDJKSA-N | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Canonical SMILES |
CC1CN(CCN1C(COC)C2=CC=C(C=C2)C(F)(F)F)C3(CCN(CC3)C(=O)C4=C(N=CN=C4C)C)C | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Isomeric SMILES |
C[C@H]1CN(CCN1[C@@H](COC)C2=CC=C(C=C2)C(F)(F)F)C3(CCN(CC3)C(=O)C4=C(N=CN=C4C)C)C | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Formula |
C28H38F3N5O2 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
DSSTOX Substance ID |
DTXSID40897719 | |
| Record name | Vicriviroc | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID40897719 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
Molecular Weight |
533.6 g/mol | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
CAS No. |
306296-47-9, 394730-30-4 | |
| Record name | Vicriviroc | |
| Source | CAS Common Chemistry | |
| URL | https://commonchemistry.cas.org/detail?cas_rn=306296-47-9 | |
| Description | CAS Common Chemistry is an open community resource for accessing chemical information. Nearly 500,000 chemical substances from CAS REGISTRY cover areas of community interest, including common and frequently regulated chemicals, and those relevant to high school and undergraduate chemistry classes. This chemical information, curated by our expert scientists, is provided in alignment with our mission as a division of the American Chemical Society. | |
| Explanation | The data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated. | |
| Record name | Vicriviroc [INN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0306296479 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | SCH-D (Old RN) | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0394730304 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Vicriviroc | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB06652 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Vicriviroc | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID40897719 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
| Record name | VICRIVIROC | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/TL515DW4QS | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
Molecular Mechanism of Action of Vicriviroc
Target Identification and Characterization: CCR5 Receptor
The C-C chemokine receptor type 5 (CCR5) is a key co-receptor for the entry of R5-tropic HIV-1 strains into target cells, primarily CD4+ T lymphocytes and macrophages. ontosight.ainih.govnih.gov R5-tropic viruses are predominantly responsible for the initial infection. frontiersin.org CCR5 is a member of the G protein-coupled receptor (GPCR) superfamily, characterized by seven transmembrane helices. nih.govdaignet.de
CCR5 Receptor Structure and Allosteric Modulation
CCR5 is a 7-transmembrane segment receptor belonging to the GPCR family. wikipedia.orgnih.govdaignet.de Small molecule CCR5 inhibitors, including vicriviroc, bind to a hydrophobic cavity located within the transmembrane domain of CCR5. nih.govnih.gov This binding site is distinct from the binding sites for natural chemokine ligands and the HIV-1 gp120 protein, indicating an allosteric mechanism of action. nih.govfrontiersin.org Allosteric modulation involves binding to a site distinct from the orthosteric site (where the natural ligand binds) and inducing a conformational change in the receptor that affects orthosteric ligand binding or receptor function. nih.govresearchgate.netnih.gov
Interaction with HIV-1 gp120 and gp41
HIV-1 entry into host cells is initiated by the binding of the viral envelope glycoprotein (Env) to the host cell's CD4 receptor. wikipedia.orgnih.govdaignet.deplos.org The Env is a trimer composed of two subunits: gp120 and gp41. daignet.deplos.orgnih.gov The gp120 subunit is responsible for binding to CD4 and the chemokine co-receptor, typically CCR5 for R5-tropic strains. wikipedia.orgnih.govdaignet.deplos.org This initial binding to CD4 induces conformational changes in gp120, exposing a high-affinity binding site for the co-receptor. daignet.deplos.org The interaction between gp120 and CCR5 then triggers further conformational changes in the gp41 subunit, which mediates the fusion of the viral membrane with the host cell membrane, allowing the viral genetic material to enter the cell. wikipedia.orgdaignet.deplos.orgnih.gov The gp41 protein undergoes a refolding process, forming a six-helix bundle that drives membrane fusion. daignet.denih.govasm.org
Binding Dynamics of this compound to CCR5
This compound exerts its inhibitory effect by binding to the CCR5 receptor and interfering with the interaction between the viral gp120 protein and the receptor. nih.govontosight.ai
Noncompetitive Allosteric Antagonism
This compound is characterized as a noncompetitive allosteric antagonist of CCR5. nih.govwikipedia.orgnih.govnih.gov This means that this compound does not compete directly with the natural chemokine ligands or the viral gp120 for the same binding site on CCR5. nih.gov Instead, it binds to a distinct allosteric site within the transmembrane domain. nih.govnih.gov Binding to this site induces a conformational change in the CCR5 receptor that prevents or significantly reduces the ability of gp120 to bind effectively to its recognition site on the receptor, thereby blocking viral entry. nih.govwikipedia.org
Specific Residue Interactions within the CCR5 Hydrophobic Pocket
This compound binds to a small hydrophobic pocket located between the transmembrane helices near the extracellular surface of the CCR5 receptor. nih.govwikipedia.org Specific interactions between this compound and residues within this pocket have been described. The trifluoromethyl phenyl group of this compound is thought to interact strongly with the I198 residue on the fifth transmembrane helix (TM5) of CCR5 through hydrophobic interactions. wikipedia.org Additionally, electrostatic interactions may occur between the positively charged tertiary nitrogen group of this compound and the hydrophilic region provided by the E283 residue on TM7 of CCR5. wikipedia.orgplos.org Other residues predicted to be involved in strong interactions include Y108 on TM3 and Y251 on TM6. wikipedia.orgplos.org Studies suggest that while this compound and maraviroc bind to a similar hydrophobic pocket, they do so in subtly different ways, interacting with distinct amino acids, which may lead to the stabilization of different CCR5 conformations. nih.gov
Predicted Key Residue Interactions of this compound with CCR5
| CCR5 Residue | Transmembrane Helix | Type of Interaction | This compound Moiety |
| I198 | TM5 | Hydrophobic | Trifluoromethyl phenyl group |
| E283 | TM7 | Electrostatic | Positively charged tertiary nitrogen |
| Y108 | TM3 | Interaction Predicted | Not specified |
| Y251 | TM6 | Interaction Predicted | Not specified |
| W86 | TM2 | Aromatic Interaction (Predicted) | Pyrimidine ring (Predicted based on similar antagonists) plos.org |
| Y37 | TM1 | Interaction Predicted | Not specified |
| F109 | TM3 | Interaction Predicted | Not specified |
| T167 | TM4 | Interaction Predicted | Not specified |
Note: Interactions are based on research findings and predictions. wikipedia.orgplos.org
Cellular and Molecular Effects of CCR5 Blockade
Blocking the CCR5 receptor with antagonists like this compound prevents the crucial interaction between the HIV-1 gp120 protein and the host cell co-receptor. nih.govontosight.aiwikipedia.org This blockade inhibits the conformational changes required in gp41 for membrane fusion to occur. daignet.deplos.orgnih.gov Consequently, the virus is unable to enter the target cell, effectively inhibiting the initial stage of the HIV-1 life cycle. nih.govontosight.aiwikipedia.org This mechanism is distinct from drugs targeting later stages of viral replication, such as reverse transcriptase or protease inhibitors. wikipedia.org By preventing viral entry, CCR5 antagonists reduce the viral load in the body. ontosight.ai
Molecular and Cellular Effects of CCR5 Blockade by this compound
| Effect | Molecular/Cellular Mechanism | Outcome |
| Inhibition of HIV-1 Entry | Prevents gp120 binding to CCR5 after CD4 interaction. nih.govontosight.aiwikipedia.org | Blocks viral fusion with host cell membrane. wikipedia.orgplos.orgnih.gov |
| Prevention of gp41 Conformational Change | Lack of gp120-CCR5 interaction inhibits the necessary structural rearrangements in gp41. daignet.deplos.orgnih.gov | Fusion machinery remains inactive. |
| Reduced Viral Load | Prevention of new cell infections. ontosight.ai | Decreased replication and spread of HIV-1. ontosight.ai |
| Inhibition of Chemokine Signaling | This compound acts as a receptor antagonist by inhibiting signaling of CCR5 by chemokines. nih.govresearchgate.net | Affects downstream cellular responses mediated by CCR5. nih.govresearchgate.net |
Note: Data is based on research findings. nih.govontosight.aiwikipedia.orgdaignet.deplos.orgnih.govnih.govresearchgate.net
Inhibition of Viral Entry
HIV-1 entry into target cells is a multi-step process initiated by the binding of the viral envelope glycoprotein gp120 to the CD4 receptor on the host cell surface. wikipedia.orghivclinic.ca This interaction induces a conformational change in gp120, exposing a binding site for a co-receptor, which is typically either CCR5 or CXCR4. wikipedia.orghivclinic.ca CCR5-tropic (R5) strains of HIV-1 utilize the CCR5 co-receptor for entry. nih.govresearchgate.netnih.gov
This compound functions as an entry inhibitor by specifically binding to the CCR5 receptor. bocsci.comnih.govresearchgate.netnih.gov It is described as a noncompetitive allosteric antagonist that binds to a hydrophobic pocket located between the transmembrane helices near the extracellular surface of the CCR5 receptor. wikipedia.orgdrugbank.comnih.gov This binding event induces a conformational change in the extracellular segment of CCR5, which in turn prevents the interaction between the viral gp120 protein and the CCR5 receptor. wikipedia.orgdrugbank.comnih.gov By blocking this crucial co-receptor binding step, this compound effectively prevents the fusion of the viral and cellular membranes, thereby inhibiting the entry of CCR5-tropic HIV-1 into target cells. wikipedia.orgontosight.aidrugbank.comnih.gov
Studies have shown that this compound potently inhibits the replication of a broad spectrum of CCR5-tropic HIV-1 isolates, including those that are genetically diverse and resistant to other classes of antiretroviral drugs. nih.govnih.gov Its antiviral activity has been demonstrated in various in vitro assays. For instance, this compound has shown potent activity against a panel of 30 R5-tropic HIV-1 isolates with EC50 values ranging from 0.04 nM to 2.3 nM. selleckchem.combiocrick.comcaymanchem.com It is also effective against viruses with defined resistance patterns to reverse transcriptase inhibitors (RTIs), protease inhibitors (PIs), or fusion inhibitors. selleckchem.combiocrick.com
Impact on Chemokine-Induced Cellular Functions
Beyond its role in viral entry, CCR5 is a G protein-coupled receptor that mediates cellular responses to its endogenous ligands, such as the chemokines MIP-1α, MIP-1β, and RANTES. nih.govdrugbank.com These interactions trigger downstream signaling pathways involved in various cellular functions, including chemotaxis, calcium flux, and GTPγS binding. nih.govresearchgate.netnih.gov As a CCR5 antagonist, this compound interferes with the binding of these chemokines, thereby inhibiting these signaling events. nih.govresearchgate.netnih.govhodoodo.com
Research findings from functional assays confirm that this compound acts as a receptor antagonist by inhibiting the signaling of CCR5 induced by chemokines. nih.govresearchgate.netnih.govhodoodo.com
Chemotaxis: this compound has been shown to inhibit chemokine-mediated migration of cells expressing human CCR5. In a chemotaxis assay using a mouse Ba/F3 cell line stably expressing recombinant human CCR5, this compound demonstrated potent inhibition of the chemotactic response induced by MIP-1α, with an IC50 value below 1 nM. nih.govresearchgate.netbiocrick.comcaymanchem.com Similar inhibitory effects were observed when RANTES and MIP-1β were used as chemoattractants. nih.gov
Calcium Flux: Stimulation of CCR5 by its ligands leads to an increase in intracellular calcium levels. This compound inhibits this intracellular calcium release induced by receptor stimulation. nih.govresearchgate.netbiocrick.com In U-87-CCR5 cells, this compound inhibited intracellular calcium release induced by RANTES with an IC50 of 16 nM. selleckchem.com Importantly, this compound treatment alone did not induce calcium signaling, confirming its antagonist properties. nih.govresearchgate.netselleckchem.com
GTPγS Binding: Receptor activation of G proteins involves the exchange of GDP for GTP, which can be measured using the binding of the non-hydrolyzable GTP analog, GTPγS. CCR5 activation by chemokines stimulates GTPγS binding to the associated G proteins. This compound inhibits this chemokine-induced GTPγS binding, indicating its ability to block downstream signaling. nih.govresearchgate.netnih.govhodoodo.com In a GTPγS binding assay, this compound potently inhibited RANTES-induced signaling with a mean IC50 of 4.2 ± 1.3 nM. nih.govselleckchem.combiocrick.com Neither this compound nor a comparator compound (SCH-C) alone induced a signal in this assay, further supporting their antagonist characteristics. nih.gov
These functional assays collectively demonstrate that this compound is a potent antagonist of the CCR5 receptor capable of inhibiting receptor function triggered by its natural chemokine ligands. nih.govresearchgate.net
Summary of In Vitro Functional Assay Results
| Assay | Stimulus | Cell Line / System | This compound IC50 (nM) | Reference |
| Chemotaxis | MIP-1α | Ba/F3-CCR5 cells | < 1 | nih.govbiocrick.comcaymanchem.com |
| Calcium Flux | RANTES | U-87-CCR5 cells | 16 | selleckchem.com |
| GTPγS Binding | RANTES | HTS-hCCR5 cell membranes | 4.2 ± 1.3 | nih.govselleckchem.combiocrick.com |
Antiviral Activity Against R5-tropic HIV-1 Isolates
| HIV-1 Isolates Panel | Geometric Mean EC50 (nM) | Geometric Mean EC90 (nM) | Reference |
| 30 R5-tropic | 0.04 - 2.3 | 0.45 - 18 | selleckchem.combiocrick.commedchemexpress.com |
Preclinical Pharmacology and in Vitro Efficacy
Antiviral Activity Spectrum
Vicriviroc has demonstrated potent and broad-spectrum antiviral activity against various HIV-1 isolates in in vitro settings. nih.govnih.gov Its mechanism of action, targeting the host cell coreceptor, is distinct from many other antiretroviral classes, which primarily target viral enzymes. natap.orgmsdmanuals.com
Activity against R5-tropic HIV-1 Isolates
This compound has shown potent inhibitory activity against R5-tropic HIV-1 isolates, which utilize the CCR5 coreceptor for entry into host cells. nih.govnih.govasm.org This is particularly relevant as R5-tropic viruses are responsible for the majority of HIV-1 transmissions and are typically the predominant species during the early stages of infection. tandfonline.comoup.com Studies using a panel of diverse R5-tropic HIV-1 isolates have shown this compound to be a potent inhibitor of viral replication. nih.govselleckchem.com
In a study evaluating the antiviral activity of this compound against 30 R5-tropic HIV-1 isolates from various genetic clades, this compound potently inhibited all tested isolates. nih.govselleckchem.com The geometric mean EC50 values ranged between 0.04 nM and 2.3 nM, while the EC90 values ranged from 0.45 nM to 18 nM. nih.govselleckchem.com this compound was consistently more active than SCH-C, an earlier CCR5 antagonist, against these isolates, showing 2- to 40-fold lower mean EC50 values. selleckchem.comresearchgate.net
| HIV-1 Isolate Source (Clade) | Geometric Mean EC50 (nM) | Geometric Mean EC90 (nM) |
| Diverse R5-tropic isolates (n=30) | 0.04 - 2.3 | 0.45 - 18 |
| Clade G Russian isolate RU570 | Not specified | 16 |
Data compiled from research findings. nih.govselleckchem.com
Activity against Drug-Resistant HIV-1 Strains
A significant challenge in HIV treatment is the emergence of drug-resistant viral strains. nih.gov Preclinical studies evaluated the activity of this compound against HIV-1 isolates resistant to existing antiretroviral drugs from other classes, such as reverse transcriptase inhibitors (RTIs) and protease inhibitors (PRIs). nih.govnatap.org
This compound demonstrated activity against viruses with RTI, PRI, or multidrug resistance (MDR) phenotypes. nih.govselleckchem.com The EC50 values against these resistant viruses were comparable to those observed for wild-type control viruses. nih.gov This finding is expected because this compound targets an early step in the viral life cycle (viral entry) that occurs prior to the targets of RTIs and PRIs (reverse transcription and virion maturation, respectively). nih.govselleckchem.com this compound also exhibited potent antiviral activity against recombinant viruses carrying multiple drug resistance mutations, including those resistant to the fusion inhibitor enfuvirtide. natap.org This suggests that resistance to other drug classes does not necessarily confer cross-resistance to this compound. natap.orgoup.com
However, resistance to CCR5 antagonists can emerge, often involving mutations in the viral envelope protein gp120, particularly in the V3 loop, or changes in coreceptor usage. tandfonline.comoup.com Studies have shown that this compound-resistant strains can exhibit cross-resistance to other CCR5 inhibitors that share similar interaction sites on the CCR5 coreceptor. oup.com
Synergistic Antiviral Effects in Combination with Other Antiretrovirals
Given that antiretroviral therapy typically involves the use of multiple drugs in combination, the potential for synergistic antiviral effects between this compound and other antiretroviral classes has been investigated in vitro. natap.orgasm.org
Mechanisms of Synergy with Different Antiretroviral Classes
In vitro drug combination studies have confirmed that this compound can act synergistically with drugs from various approved antiretroviral classes. nih.govnih.govnatap.orgasm.org Synergy has been observed with nucleoside reverse transcriptase inhibitors (NRTIs) such as zidovudine and lamivudine, the non-nucleoside reverse transcriptase inhibitor (NNRTI) efavirenz, and the protease inhibitor indinavir. asm.org Additionally, this compound has shown synergy with the fusion inhibitor enfuvirtide, indicating that simultaneously targeting multiple steps in the viral entry process can lead to cooperative effects. asm.org
The mechanisms underlying the synergy between this compound and other antiretrovirals are related to their distinct targets in the HIV-1 life cycle. By blocking viral entry at the CCR5 coreceptor, this compound reduces the number of target cells infected, while drugs from other classes inhibit subsequent steps like reverse transcription, protein processing, or viral fusion. natap.orgmsdmanuals.com This multi-targeted approach can lead to enhanced viral inhibition compared to the use of single agents. tandfonline.com
Synergy has also been reported between this compound and other CCR5 inhibitors, such as the monoclonal antibody PRO 140 and maraviroc, suggesting that targeting different sites or inducing distinct conformational changes on the CCR5 receptor can result in synergistic antiviral effects. asm.orgresearchgate.net Competition binding studies support the idea that these agents may interact with CCR5 in different ways. asm.org
Furthermore, studies have shown that reducing CCR5 density on target cells with agents like rapamycin can enhance the antiviral activity of this compound, including against this compound-resistant strains, suggesting a potential synergistic interaction through modulation of the host cell environment. pnas.orgnih.gov
Receptor Binding and Functional Assays
Preclinical studies have characterized the interaction of this compound with the CCR5 receptor through binding and functional assays to understand its mechanism as a CCR5 antagonist. nih.govnih.gov
Competitive Binding Assays
Competition binding assays are used to determine the affinity of a compound for its target receptor. nih.govnih.gov These assays have shown that this compound binds specifically to the CCR5 receptor. nih.govnih.gov
In competition binding assays, this compound demonstrated a higher binding affinity for CCR5 compared to SCH-C. nih.govnih.govselleckchem.comresearchgate.net The reported Ki value for this compound binding to CCR5 is 0.8 nM, while that for SCH-C is 2.6 nM. selleckchem.com This higher affinity is thought to contribute to this compound's greater antiviral potency observed in cell-based assays. selleckchem.comresearchgate.net
Binding kinetics studies have also shown that this compound dissociates from CCR5 at a slower rate than maraviroc, another CCR5 antagonist, which may offer a pharmacodynamic advantage in maintaining sustained receptor occupancy. natap.org
Functional assays, including the inhibition of calcium flux, guanosine 5′-[35S]triphosphate (GTPγS) exchange, and chemotaxis, have further confirmed that this compound acts as a receptor antagonist by inhibiting the signaling of CCR5 by chemokines like RANTES. nih.govnih.govselleckchem.comresearchgate.net this compound inhibits RANTES-induced intracellular calcium release and GTPγS binding to CCR5 membranes, demonstrating its ability to block downstream signaling events mediated by the receptor. selleckchem.comresearchgate.net
Inhibition of CCR5-Mediated Signaling Pathways
This compound acts as a potent antagonist of the CCR5 receptor, effectively inhibiting its function. nih.gov Studies utilizing functional assays such as calcium flux, chemotaxis, and GTPγS exchange have demonstrated its ability to block signaling mediated by chemokines that bind to CCR5. nih.gov
In chemotaxis assays using a mouse Ba/F3 cell line stably expressing human CCR5, this compound demonstrated potent inhibition of the chemotactic response induced by MIP-1α, with IC50 values below 1 nM. nih.gov Similar inhibitory effects were observed when RANTES and MIP-1β were used to induce chemotaxis. nih.gov
In U-87-CCR5 cells, this compound inhibited intracellular calcium release triggered by the ligand RANTES, with an IC50 of 16 nM. selleckchem.com Importantly, this compound treatment alone did not stimulate calcium release in this assay, confirming its antagonist properties. nih.govselleckchem.com
The ability of this compound to inhibit CCR5 receptor signaling was further demonstrated in a GTPγS exchange assay using membranes from HTS-hCCR5 cells. nih.gov this compound potently inhibited RANTES-induced GTPγS binding, with a mean IC50 of 4.2 ± 1.3 nM. nih.govselleckchem.com Neither this compound nor a related compound, SCH-C, induced a signal in this assay, further supporting their antagonist nature. nih.gov
This compound has also been shown to block CCL5-activated signaling mediated by CCR5 in basal-like human breast cancer cells, with IC50 values reported below 30 nM. aacrjournals.org
The following table summarizes some of the in vitro efficacy data regarding the inhibition of CCR5-mediated signaling pathways:
| Assay | Cell Line / System | Ligand | IC50 (nM) | Reference |
| Chemotaxis | Ba/F3-CCR5 cells | MIP-1α | < 1 | nih.gov |
| Intracellular Calcium Release | U-87-CCR5 cells | RANTES | 16 | selleckchem.com |
| GTPγS Binding | HTS-hCCR5 cell membranes | RANTES | 4.2 ± 1.3 | nih.govselleckchem.com |
| CCL5-activated signaling | Basal-like human breast cancer cells | CCL5 | < 30 | aacrjournals.org |
Preclinical Pharmacokinetic Considerations (in vitro metabolism)
Preclinical Pharmacokinetic Considerations (in vitro metabolism)
Metabolic Pathways and Enzyme Involvement (e.g., CYP3A4, CYP3A5, CYP2C9)
In vitro metabolism studies using human liver microsomes have been conducted to identify the cytochrome P450 (CYP) enzymes responsible for the biotransformation of this compound. nih.govresearchgate.net These studies indicate that this compound is metabolized via several oxidative pathways, including N-oxidation, O-demethylation, N,N-dealkylation, N-dealkylation, and oxidation to a carboxylic acid metabolite. nih.govresearchgate.net
The primary enzyme responsible for the formation of the major this compound metabolites in human liver microsomes is CYP3A4. nih.gov Recombinant human CYP3A4 has been shown to catalyze the formation of all identified metabolites. nih.govresearchgate.net
CYP3A5 and CYP2C9 also play roles in this compound biotransformation, although they are considered minor contributors compared to CYP3A4. nih.govresearchgate.net Recombinant human CYP3A5 catalyzes the formation of N-oxidation (M2/M3) and N-dealkylation (M41) metabolites, while CYP2C9 primarily catalyzes the formation of the O-demethylation metabolite (M15). nih.gov
Studies using CYP3A4 inhibitors like ketoconazole and azamulin have demonstrated potent inhibition of the formation of multiple this compound metabolites in human liver microsomes. nih.gov A CYP3A4/5-specific monoclonal antibody also significantly inhibited the formation of all metabolites, by 86% to 100%. nih.gov
Protein Binding Characteristics
Plasma protein binding is an important consideration in pharmacokinetics as only unbound drug is generally considered free to exert therapeutic effects, be metabolized, or be excreted. bioivt.comsygnaturediscovery.com
Studies evaluating the binding of this compound to human plasma proteins have shown it to be moderately bound. nih.gov Approximately 16% of the compound remained unbound in human plasma when evaluated at a concentration of 0.94 µM using ultrafiltration. nih.govresearchgate.net
In vitro studies performed with increasing amounts of human serum showed no significant effect of serum concentration on the antiviral activity of this compound at concentrations up to 50% in the cultures. nih.gov Based on these findings, it was not anticipated that serum protein binding would have a significant impact on the in vivo antiviral efficacy of this compound. nih.gov
Mechanisms of Hiv-1 Resistance to Vicriviroc
Molecular Basis of Resistance Development
The development of resistance to vicriviroc at the molecular level is often associated with changes in the HIV-1 envelope protein, particularly in gp120. These changes can affect the interaction between gp120 and the CCR5 coreceptor, even in the presence of the inhibitor.
Mutations in HIV-1 gp120 V3 Loop Region
The V3 loop of HIV-1 gp120 is a critical determinant of coreceptor usage and a primary site for mutations conferring resistance to CCR5 antagonists like this compound. mdpi.com Studies have shown that amino acid substitutions within the V3 loop are frequently associated with reduced susceptibility to this compound. oup.comasm.orgnih.govnih.govasm.org These mutations can arise sequentially, leading to a complete loss of drug activity. nih.gov
Specific mutations in the V3 loop have been identified in this compound-resistant viruses, although a consistent, conserved pattern across all subjects is not always observed. oup.comnih.gov Examples of V3 loop mutations associated with this compound resistance include Q18E, D25G, and G17A, which were observed in one study and linked to progressive reductions in susceptibility. oup.com The K10R substitution has also been noted in some this compound-resistant viruses. oup.com In a study of an HIV-1 subtype D isolate, V3 loop mutations Q315E and R321G were found to be essential for resistance. nih.gov
However, the impact of V3 loop mutations on this compound susceptibility can depend on the context of the entire envelope glycoprotein sequence. nih.gov Viruses with identical V3 loop sequences have shown varying degrees of susceptibility, indicating that other regions of gp160 also play a role. oup.com
Mutations Outside the V3 Loop (e.g., C4)
While the V3 loop is a major contributor to this compound resistance, mutations in other regions of gp120 and even gp41 can also play a significant role. oup.comnih.govoup.com Substitutions outside the V3 loop, such as in the C4 domain of gp120 and the fusion peptide of gp41, have been shown to contribute to the resistance phenotype. oup.comnih.govoup.com
For instance, in the study involving the HIV-1 subtype D isolate, in addition to V3 loop mutations, changes in the C4 domain, specifically E328K and G429R, contributed significantly to the infectivity of the resistant variant. nih.gov These mutations outside the V3 loop can modulate the magnitude of resistance. researchgate.net Studies have indicated that mutations both within and outside the V3 loop contribute to the phenotypic resistance to this compound, and there is no clearly conserved pattern of mutations in gp160 associated with this resistance. nih.gov
Adaptation to Drug-Bound CCR5 Conformation
A key mechanism of resistance to CCR5 antagonists like this compound involves the virus adapting to utilize the drug-bound conformation of the CCR5 coreceptor for entry. oup.comasm.orgnih.govnih.govresearchgate.net this compound, similar to other CCR5 antagonists, binds to a hydrophobic pocket within the transmembrane domains of CCR5, inducing conformational changes in the receptor. oup.comasm.orgnih.govasm.org Resistant viruses develop alterations in their envelope protein that allow them to interact effectively with this altered, drug-bound state of CCR5. oup.comasm.org
This adaptation can manifest phenotypically as a reduction in the maximum percent inhibition (MPI) of viral entry by the drug, rather than a significant increase in the half-maximal inhibitory concentration (IC50). asm.orgresearchgate.neti-base.info This suggests the virus is not simply requiring higher drug concentrations to block entry via unbound CCR5, but is instead gaining the ability to enter via the drug-occupied receptor. asm.orgresearchgate.net The ability to use the drug-bound CCR5 conformation is suggested to correlate with a reduction in MPI. researchgate.netasm.org
Studies have shown that this compound-resistant viruses can exhibit enhanced replication in the presence of the drug, indicating successful adaptation to using the drug-bound CCR5. nih.govnih.gov This adaptation often involves increased reliance on specific regions of CCR5, such as the N-terminus. nih.govnih.govresearchgate.net
Viral Tropism Switching as an Escape Mechanism
Another significant mechanism of escape from CCR5 antagonists is a shift in viral tropism. While this compound is designed to inhibit viruses that use the CCR5 coreceptor (R5-tropic viruses), resistance can emerge through the selection or evolution of viruses capable of using the CXCR4 coreceptor (X4-tropic) or both CCR5 and CXCR4 (dual/mixed-tropic). mdpi.comoup.comnih.govnih.gov
Emergence of X4-tropic or Dual/Mixed-tropic Viruses
The emergence of X4-tropic or dual/mixed-tropic viruses is a well-documented escape pathway from CCR5 antagonist therapy. mdpi.comoup.comresearchgate.netnih.govnih.govnatap.orgi-base.infoamanote.com These viruses are not inhibited by this compound because they utilize CXCR4 for entry, either exclusively or in addition to CCR5. natap.org In clinical trials of this compound, the emergence of dual/mixed-tropic or X4 viruses was observed in a proportion of subjects experiencing virological failure. oup.comnih.govnatap.orgi-base.infonatap.orgaidsmap.com
The prevalence of tropism switching can vary, and in some studies, it was not the dominant mechanism of failure. oup.comnih.gov However, the presence of pre-existing minor populations of X4-using variants at baseline can be a significant factor in the emergence of these viruses under CCR5 antagonist pressure. portlandpress.com Studies have shown that even small populations of non-R5 strains present before treatment can be selected for when the R5 population is suppressed by the drug. portlandpress.com
Data from clinical trials indicate that the emergence of dual/mixed or X4 virus can occur relatively early during treatment and does not always coincide precisely with virological failure. aidsmap.com The likelihood of tropism switch might also be influenced by the this compound dose. natap.orgi-base.info
Genetic Determinants of Tropism Shift
The shift in viral tropism from CCR5 to CXCR4 usage is primarily determined by changes in the HIV-1 envelope protein, particularly within the V3 loop of gp120. mdpi.comnih.gov The V3 loop sequence is a key predictor of coreceptor usage. mdpi.com
Mutations in the V3 loop can alter the coreceptor specificity of the virus, enabling it to utilize CXCR4. mdpi.com As few as two amino acid changes in the V3 loop have been shown to change coreceptor specificity from CCR5 to CXCR4. mdpi.com While the V3 loop is the primary determinant, other regions of gp120, such as V1 and V2, and even C2, have also been implicated in influencing coreceptor usage and tropism shifts. mdpi.comresearchgate.netnih.gov
Genotypic analysis of viruses that undergo tropism switching often reveals specific amino acid changes in the V3 loop and potentially other envelope regions that are associated with the ability to use CXCR4. oup.comnih.govresearchgate.net These genetic determinants facilitate the virus's ability to bind to and enter cells via the CXCR4 coreceptor, providing an escape route from this compound inhibition.
Table 1: Summary of Mechanisms of this compound Resistance
| Mechanism | Description | Key Viral Component(s) Involved | Examples of Genetic Changes | Phenotypic Manifestation |
| Molecular Basis of Resistance | Viral adaptation to enter via drug-bound CCR5. | gp120 (V3 loop, C4), gp41 | Mutations in V3 loop (e.g., Q18E, D25G, G17A, K10R, Q315E, R321G), mutations in C4 (e.g., E328K, G429R), mutations in gp41. oup.comnih.govnih.gov | Reduced Maximum Percent Inhibition (MPI). asm.orgresearchgate.neti-base.info |
| Viral Tropism Switching | Shift from CCR5 usage to CXCR4 or dual/mixed usage. | gp120 (V3 loop, V1/V2, C2) | Mutations in V3 loop primarily, also in V1/V2 and C2. mdpi.comresearchgate.netnih.gov | Emergence of X4-tropic or Dual/Mixed-tropic viruses. oup.comnih.govresearchgate.netnatap.orgi-base.infoamanote.com |
Table 2: Examples of V3 Loop Mutations Associated with this compound Resistance
| Mutation | Location (HXB2 numbering) | Associated with | Reference |
| Q18E | V3 loop | Reduced this compound susceptibility | oup.com |
| D25G | V3 loop | Reduced this compound susceptibility | oup.com |
| G17A | V3 loop | Further reductions in this compound susceptibility | oup.com |
| K10R | V3 loop | Observed in some resistant viruses | oup.com |
| Q315E | V3 loop | Essential for resistance (Subtype D isolate) | nih.gov |
| R321G | V3 loop | Essential for resistance (Subtype D isolate) | nih.gov |
| S306P | V3 loop | Associated with complete phenotypic resistance | nih.govasm.org |
Cross-Resistance Profiles with Other CCR5 Antagonists
Resistance to one CCR5 antagonist does not always translate to resistance to others in the same class. oup.com The cross-resistance profile of this compound with other CCR5 antagonists is complex and depends on the specific viral isolate and the mutations that emerge.
Comparison with Maraviroc and Other Agents (e.g., TAK779, SCH-C, Aplaviroc)
This compound and maraviroc are both small-molecule CCR5 antagonists that bind to similar sites on the CCR5 receptor, inducing conformational changes that prevent gp120 binding. asm.orgwikipedia.orgnih.gov However, their binding interactions can be subtly different, leading to varied cross-resistance profiles. researchgate.netnatap.org
Studies have shown that viruses resistant to maraviroc can remain sensitive to this compound and aplaviroc. researchgate.netnih.govoup.com Conversely, this compound-resistant strains have demonstrated cross-resistance to TAK779 and SCH-C, suggesting shared interaction sites or similar allosteric changes induced in the CCR5 receptor by these compounds. asm.orgnih.govoup.comasm.org For instance, a this compound-resistant virus from a subject in a clinical study showed cross-resistance to TAK779. asm.orgnih.gov
The mechanism of resistance, specifically whether the virus primarily utilizes the N-terminus or requires both the N-terminus and extracellular loops (ECLs) of CCR5 for entry in the presence of the drug, appears to influence the breadth of cross-resistance. researchgate.netnih.gov Viruses that predominantly utilize the N-terminus may exhibit broad cross-resistance, while those requiring both N-terminus and antagonist-specific ECL changes might show a narrower cross-resistance profile. researchgate.netnih.gov
Some studies have indicated that this compound-resistant viruses may not have amino acid changes in the V3 region, unlike resistance to other CCR5 antagonists like SCH-C. asm.org However, other research highlights the importance of V3 loop mutations in conferring this compound resistance. asm.orgnih.govcore.ac.uki-base.info The specific mutations associated with resistance can vary depending on the HIV-1 subtype and the environmental context. asm.orgcore.ac.uk
The following table summarizes some observed cross-resistance patterns among CCR5 antagonists:
| CCR5 Antagonist Resistance | Observed Cross-Resistance To | Observed Sensitivity To | Source |
| This compound | TAK779, SCH-C, AD101 | Maraviroc, Aplaviroc (in some cases) | asm.orgnih.govoup.comoup.comasm.org |
| Maraviroc | TAK779 (partial in some cases) | This compound, Aplaviroc, SCH-C | researchgate.netnih.govoup.comasm.org |
| Aplaviroc | AD101, TAK779, SCH-C, Maraviroc (in one specific case) | asm.orgduke.edu | |
| TAK-652 | TAK-779 (partial) | TAK-220, Maraviroc, this compound, Aplaviroc | asm.org |
Note: This table reflects observed patterns in specific studies and may not be exhaustive or universally applicable to all viral isolates.
Impact of Resistance on Viral Fitness (in vitro)
The impact of developing resistance to CCR5 antagonists on viral fitness in vitro has been investigated, with varying findings depending on the specific antagonist and viral isolate studied. Viral fitness is generally defined as the efficiency of viral replication. portlandpress.com
Some in vitro data have suggested that resistance to certain CCR5 antagonists, such as AD101 (a precursor of this compound), may not confer a significant loss of viral fitness. asm.orgnih.govresearchgate.netplos.org However, other studies, particularly those examining the decay of resistance mutations after drug discontinuation in vivo, imply that this compound resistance may be associated with a fitness cost. asm.orgnih.govresearchgate.netnih.gov
Specifically, the loss of this compound resistance-associated mutations in the V3 loop after the discontinuation of the drug in a clinical setting suggests that these resistant variants may have a reduced replicative capacity in the absence of drug pressure compared to wild-type virus. asm.orgnih.govresearchgate.netnih.gov One study estimated a fitness disadvantage of 10% to 11% for this compound-resistant viruses in vivo in the absence of the drug. nih.gov This contrasts with some in vitro findings where resistance to a related compound (AD101) was not associated with a fitness loss. asm.orgnih.govresearchgate.netplos.org
The development of resistance through the use of drug-bound CCR5 may involve complex alterations in how the viral envelope protein engages the receptor, which could potentially impact viral entry efficiency and, consequently, fitness. ingentaconnect.com While resistant viruses are more fit in the presence of the selecting drug, the fitness cost in the absence of the drug is a crucial factor influencing the persistence of resistant variants. portlandpress.com
Studies on maraviroc resistance in vitro have also explored the impact on fitness, with some findings suggesting that while resistance emerges, the viruses maintain their replicative capacity. researchgate.net However, the specific mutations and mechanisms of resistance can influence the fitness landscape.
Computational and Structural Biology Approaches in Vicriviroc Research
Molecular Modeling of CCR5 and Vicriviroc Interactions
Molecular modeling techniques are essential for visualizing and analyzing the complex interactions between small molecules like this compound and a transmembrane protein like CCR5. Given the challenges in obtaining high-resolution experimental structures of G protein-coupled receptors (GPCRs) like CCR5, computational modeling provides valuable structural information.
Homology Modeling of CCR5 Receptor Structure
Homology modeling has been widely used to generate three-dimensional structures of the CCR5 receptor. This technique relies on the known experimental structures of homologous proteins, such as other chemokine receptors or rhodopsin, as templates. Early models of CCR5 were often based on the bovine rhodopsin structure. plos.orgnih.gov More recent studies have utilized templates with higher sequence identity and resolution, such as the crystal structure of CXCR4 (PDB code: 3ODU). plos.orgnih.govmdpi.comnatap.org These homology models provide a structural framework for understanding the arrangement of the transmembrane helices and extracellular loops that form the binding pocket.
Docking Studies of this compound into CCR5 Binding Pocket
Molecular docking studies are employed to predict the preferred orientation and binding pose of this compound within the CCR5 binding pocket. These studies aim to determine the key residues involved in the interaction and the types of forces that stabilize the complex. This compound is known to bind to a hydrophobic pocket located between the transmembrane helices near the extracellular surface of the CCR5 receptor. wikipedia.orgasm.orgnih.gov
Docking simulations have predicted specific interactions between this compound and residues within this pocket. For instance, the trifluoromethyl phenyl group of this compound may engage in hydrophobic interactions with residues like I198 on transmembrane helix 5 (TM5). wikipedia.org Electrostatic interactions have also been predicted, such as a salt bridge between the positively charged tertiary nitrogen of this compound and the hydrophilic region containing E283 on TM7. plos.orgwikipedia.orgcapes.gov.br Other residues potentially involved in this compound binding include W86 (TM2), Y251 (TM6), and possibly Y108 (TM3), although interactions with Y108 may differ compared to other CCR5 antagonists like maraviroc. asm.org Docking studies with this compound and other CCR5 antagonists like maraviroc, SCH-C, and TAK779 have helped to delineate the nature of the CCR5 binding pocket and the unique interaction profiles of different antagonists. plos.orgcapes.gov.br
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations are used to study the dynamic behavior of the CCR5-vicriviroc complex over time. These simulations provide insights beyond static docking poses, allowing researchers to assess the stability of the binding complex, the conformational changes induced in the receptor upon ligand binding, and the flexibility of the binding pocket. nih.govmdpi.comresearchgate.netscirp.orgrsc.orgtandfonline.commdpi.comnih.gov MD simulations can refine homology models and provide a more realistic representation of the protein-ligand interactions in a membrane environment. nih.govoup.com Analysis of MD trajectories can reveal the persistence of key interactions identified in docking studies and identify additional transient interactions that contribute to binding affinity and receptor modulation. rsc.orgtandfonline.com
Structure-Based Drug Design for Next-Generation CCR5 Antagonists
Understanding the detailed molecular interactions between this compound and CCR5 through structural and computational methods is fundamental to structure-based drug design (SBDD) efforts aimed at developing next-generation CCR5 antagonists. plos.orgcapes.gov.brscirp.orgtandfonline.comosti.govresearchgate.net By providing insights into the structural features of the binding pocket and the crucial interactions required for potent antagonism, these studies guide the design of novel compounds with improved properties. SBDD approaches leverage the information from homology models, docking studies, and molecular dynamics simulations to design molecules with optimized shapes, sizes, and chemical functionalities to enhance binding affinity, specificity, and pharmacokinetic profiles, while potentially mitigating issues like toxicity and resistance seen with earlier compounds. capes.gov.brscirp.orgosti.govresearchgate.net
Computational Prediction of Resistance Pathways
Computational methods are valuable tools for investigating and predicting the mechanisms by which HIV-1 develops resistance to CCR5 antagonists like this compound. Resistance often involves mutations in the viral envelope protein gp120, particularly in the V3 loop, which allow the virus to utilize the drug-bound conformation of CCR5 for entry. nih.govnih.govasm.org Computational analyses, including evolutionary modeling and sequence analysis, can help identify key mutations associated with resistance and understand how these mutations alter the interaction between gp120 and the modified CCR5 receptor. asm.orgresearchgate.net Predicting potential resistance pathways computationally can inform the design of new inhibitors that are less susceptible to the development of resistance.
Future Directions and Broader Implications for Antiretroviral Research
Insights from Vicriviroc Research for Novel Entry Inhibitors
This compound's mechanism of action involves binding to a hydrophobic pocket within the transmembrane helices of the CCR5 receptor, inducing a conformational change that prevents the binding of the viral envelope protein gp120 and subsequent entry of HIV-1 into the host cell wikipedia.orgdrugbank.com. This allosteric antagonism of a host co-receptor represented a novel strategy compared to earlier antiretrovirals that primarily targeted viral enzymes wikipedia.org.
Research with this compound demonstrated potent antiviral activity against CCR5-tropic HIV-1 isolates in vitro and showed significant reductions in viral load in treatment-experienced patients when used in combination with an optimized background regimen in Phase 2 studies nih.govoup.comdaigonline.de. For instance, in the ACTG 5211 study, this compound demonstrated a significantly greater decline in HIV-RNA compared to placebo after 24 weeks in treatment-experienced subjects with virologic failure daigonline.de. Extended follow-up of higher dosage arms in this study showed long-term antiviral activity daigonline.de.
The studies with this compound, alongside other CCR5 antagonists like maraviroc and aplaviroc, underscored the principle that blocking host cell co-receptors is a viable strategy for inhibiting HIV-1 entry nih.govdaig-net.de. The observation that this compound could synergize with the fusion inhibitor enfuvirtide suggested that targeting multiple steps in the entry process could yield cooperative effects, potentially by prolonging the exposure of intermediate viral conformations targeted by fusion inhibitors nih.gov. This provided a rationale for exploring combinations of different classes of entry inhibitors.
Data from clinical trials, such as VICTOR-E1, showed virologic response rates in treatment-experienced individuals receiving this compound with an optimized background regimen. For example, in VICTOR-E1, 59% of patients receiving 30 mg of this compound achieved a viral load below 50 copies/mL at week 48, compared to 25% in the placebo group natap.org. These findings, while part of a development program that ultimately did not lead to approval, contributed to the understanding of the potential efficacy of CCR5 antagonists in specific patient populations.
Addressing Challenges in CCR5 Antagonist Development
The development of this compound encountered several challenges that have informed subsequent CCR5 antagonist research. One significant challenge is the necessity of identifying the tropism of the infecting HIV-1 strain, as CCR5 antagonists are only effective against CCR5-tropic virus wikipedia.orgnih.gov. The prevalence of CXCR4-using or dual/mixed-tropic virus can impact treatment outcomes wikipedia.orgoup.com. Studies with this compound showed diminished virologic and immunologic responses in subjects with dual/mixed viruses at entry compared to those with R5 virus, although responses were still better than in placebo recipients oup.com. This highlights the critical need for accurate and accessible tropism testing prior to initiating CCR5 antagonist therapy nih.gov.
Another challenge observed in the development of CCR5 antagonists, including this compound, was the potential for viral breakthrough and the emergence of resistance. While reports suggest that coreceptor switch is a rare cause of viral escape under CCR5 antagonist treatment, the possibility exists oup.com. Studies with this compound also faced challenges related to demonstrating non-inferiority compared to existing therapies in treatment-naïve patients, leading to the termination of some studies in this population aidsmap.comdaig-net.de.
The experience with this compound and other CCR5 antagonists also raised considerations regarding potential off-target effects due to the role of CCR5 in normal immune function oup.com. Although concerns about potential class-specific long-term adverse effects like hepatotoxicity or malignancy were raised, pooled data from some studies did not show a new or added toxicity risk for this compound compared to placebo arms at the time of analysis daigonline.deoup.com. However, the discontinuation of aplaviroc due to hepatotoxicity underscored the importance of vigilant safety monitoring in this drug class oup.comfrontiersin.org.
Continued Relevance of Host-Targeting Strategies in Antiviral Research
The research surrounding this compound reinforces the continued relevance of host-targeting strategies in antiviral research. Unlike traditional antivirals that target viral components, host-targeted antivirals (HTAs) interfere with host cell factors or pathways that viruses utilize for replication nih.govlongdom.org. This approach offers potential advantages, including a higher genetic barrier to resistance because host targets are generally less prone to rapid mutation than viral proteins, and the potential for broad-spectrum activity against different viruses that rely on similar host pathways asm.orgresearchgate.netfrontiersin.org.
The development of CCR5 antagonists like this compound was a pioneering effort in the field of HTAs for HIV. Although this compound did not achieve widespread clinical use, the knowledge gained from its research and clinical trials has been invaluable. It provided crucial data on the feasibility and challenges of targeting a host co-receptor for antiviral therapy.
The concept of targeting host factors is gaining increasing attention for a range of viral infections researchgate.netlongdom.org. Research is exploring HTAs that interfere with various stages of the viral life cycle, including entry, biosynthesis, and release nih.gov. The experience with this compound contributes to the broader understanding of how to identify and validate host targets, the complexities of developing drugs that modulate host pathways, and the importance of carefully assessing both efficacy and potential host-mediated side effects. The ongoing research into host-targeting antivirals for various pathogens demonstrates the enduring impact of the insights gleaned from the development of compounds like this compound researchgate.netcardiff.ac.uk.
Q & A
Q. What experimental models are most appropriate for evaluating Vicriviroc’s mechanism of action as a CCR5 antagonist?
this compound’s mechanism involves blocking HIV-1 entry via CCR5 co-receptor antagonism. In vitro models using CCR5-tropic HIV-1 isolates in peripheral blood mononuclear cells (PBMCs) or cell lines like HEK293/CCR5 are standard for assessing antiviral activity . In vivo models include humanized mice or non-human primates infected with CCR5-tropic HIV-1. Dose-response curves and IC50 values should be calculated using validated assays (e.g., p24 antigen reduction assays) .
Q. How do researchers reconcile discrepancies between tropism assay results (original vs. enhanced-sensitivity) in this compound clinical trials?
Original tropism assays may misclassify dual/mixed (DM) tropic viruses as CCR5-tropic (R5), leading to suboptimal virologic responses. Enhanced-sensitivity assays (e.g., population-based sequencing with deep sequencing) improve detection of minority X4/DM variants. Researchers should retest baseline samples with enhanced assays and stratify outcomes based on updated tropism classifications .
Q. What pharmacokinetic (PK) parameters are critical for optimizing this compound dosing in clinical studies?
Key PK parameters include:
- Ctrough : Maintain concentrations above the protein-binding-adjusted IC90 (e.g., 30 mg daily achieves ~50 ng/mL trough levels) .
- Half-life : this compound’s half-life (~24–30 hours) supports once-daily dosing .
- Drug-drug interactions : Co-administration with ritonavir-boosted protease inhibitors (PI/r) increases this compound exposure by ~2-fold, necessitating dose adjustments .
Advanced Research Questions
Q. How should clinical trials be designed to address this compound’s variable efficacy across doses (e.g., 5 mg vs. 15 mg) in treatment-experienced patients?
Phase II trials (e.g., ACTG A5211) used a double-blind, randomized design with dose escalation (5–30 mg) and optimized background regimens (OBR). Key considerations:
- Primary endpoint : Change in HIV-1 RNA at day 14 (short-term activity) .
- Secondary endpoints : CD4 count changes, safety/tolerability at 24–48 weeks .
- Dose selection : Higher doses (15–30 mg) showed sustained virologic suppression, while lower doses (5 mg) had higher failure rates due to emergent X4 tropism .
| Dose (mg) | Mean HIV-1 RNA Reduction (log10 copies/mL) at 24 Weeks | Virologic Failure Rate (%) |
|---|---|---|
| 5 | -1.51 | 40 |
| 10 | -1.86 | 27 |
| 15 | -1.68 | 33 |
| Data from ACTG A5211 trial |
Q. What methodologies identify this compound resistance in patients with virologic failure despite CCR5-tropic virus?
Resistance mechanisms include:
- Phenotypic assays : Reduced maximal percentage inhibition (MPI) in viral entry assays .
- Genotypic analysis : Env gp160 mutations (e.g., V3 loop changes like G316E/K), though no consistent pattern across subjects .
- Coreceptor switching : Longitudinal monitoring for X4/DM variants via enhanced tropism assays .
Q. How do researchers interpret conflicting data on this compound’s long-term safety, including malignancy risks?
In ACTG A5211, malignancies occurred in 6 this compound recipients vs. 2 placebo recipients, but causality was unclear due to small sample sizes and confounding factors (e.g., advanced HIV disease) . Mitigation strategies:
- Extended follow-up : Monitor malignancy rates in open-label extensions (median follow-up >40 weeks) .
- Comparative safety trials : Use matched cohorts in Phase III studies (e.g., VICTOR-E1) to assess risk-benefit ratios .
Data Analysis & Contradictions
Q. How should researchers address contradictions in this compound’s dose-response relationships across trials?
Conflicting efficacy data (e.g., 15 mg underperforming vs. 10 mg in some studies) may stem from:
- OBR variability : Differential OBR potency across cohorts .
- Tropism assay sensitivity : Enhanced assays reclassify responders/non-responders .
- PK variability : Suboptimal trough levels in certain populations (e.g., CYP3A4 fast metabolizers) . Solutions include stratified analysis by OBR quality and PK-guided dosing.
Q. What statistical approaches are recommended for analyzing this compound’s efficacy in heterogeneous patient populations?
Use mixed-effects models to account for:
Featured Recommendations
| Most viewed | ||
|---|---|---|
| Most popular with customers |
Disclaimer and Information on In-Vitro Research Products
Please be aware that all articles and product information presented on BenchChem are intended solely for informational purposes. The products available for purchase on BenchChem are specifically designed for in-vitro studies, which are conducted outside of living organisms. In-vitro studies, derived from the Latin term "in glass," involve experiments performed in controlled laboratory settings using cells or tissues. It is important to note that these products are not categorized as medicines or drugs, and they have not received approval from the FDA for the prevention, treatment, or cure of any medical condition, ailment, or disease. We must emphasize that any form of bodily introduction of these products into humans or animals is strictly prohibited by law. It is essential to adhere to these guidelines to ensure compliance with legal and ethical standards in research and experimentation.


