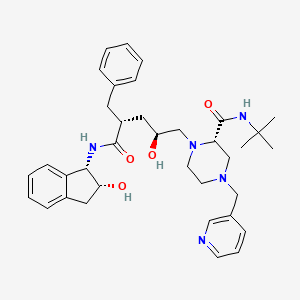
Indinavir
Description
Indinavir is a potent human immunodeficiency virus (HIV) protease inhibitor that competitively binds to the viral protease’s active site, preventing the cleavage of viral polyproteins essential for maturation and infectivity . It is metabolized primarily by cytochrome P450 3A4 (CYP3A4), with significant hepatic first-pass metabolism contributing to variable bioavailability across species . This compound is clinically used in combination with ritonavir (a CYP3A4 inhibitor) to enhance pharmacokinetics, forming a cornerstone of antiretroviral therapy (ART) for HIV-1 .
Propriétés
IUPAC Name |
(2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-5-[[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]amino]-5-oxopentyl]-N-tert-butyl-4-(pyridin-3-ylmethyl)piperazine-2-carboxamide | |
|---|---|---|
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI |
InChI=1S/C36H47N5O4/c1-36(2,3)39-35(45)31-24-40(22-26-12-9-15-37-21-26)16-17-41(31)23-29(42)19-28(18-25-10-5-4-6-11-25)34(44)38-33-30-14-8-7-13-27(30)20-32(33)43/h4-15,21,28-29,31-33,42-43H,16-20,22-24H2,1-3H3,(H,38,44)(H,39,45)/t28-,29+,31+,32-,33+/m1/s1 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI Key |
CBVCZFGXHXORBI-PXQQMZJSSA-N | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Canonical SMILES |
CC(C)(C)NC(=O)C1CN(CCN1CC(CC(CC2=CC=CC=C2)C(=O)NC3C(CC4=CC=CC=C34)O)O)CC5=CN=CC=C5 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Isomeric SMILES |
CC(C)(C)NC(=O)[C@@H]1CN(CCN1C[C@H](C[C@@H](CC2=CC=CC=C2)C(=O)N[C@@H]3[C@@H](CC4=CC=CC=C34)O)O)CC5=CN=CC=C5 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Formula |
C36H47N5O4 | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Related CAS |
157810-81-6 (sulfate (1:1) (salt)) | |
| Record name | Indinavir [USAN:INN:BAN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0150378179 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
DSSTOX Substance ID |
DTXSID4043802 | |
| Record name | Indinavir | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID4043802 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
Molecular Weight |
613.8 g/mol | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Physical Description |
Solid | |
| Record name | Indinavir | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014369 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Solubility |
4.82e-02 g/L | |
| Record name | Indinavir | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00224 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Indinavir | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014369 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
CAS No. |
150378-17-9 | |
| Record name | Indinavir | |
| Source | CAS Common Chemistry | |
| URL | https://commonchemistry.cas.org/detail?cas_rn=150378-17-9 | |
| Description | CAS Common Chemistry is an open community resource for accessing chemical information. Nearly 500,000 chemical substances from CAS REGISTRY cover areas of community interest, including common and frequently regulated chemicals, and those relevant to high school and undergraduate chemistry classes. This chemical information, curated by our expert scientists, is provided in alignment with our mission as a division of the American Chemical Society. | |
| Explanation | The data from CAS Common Chemistry is provided under a CC-BY-NC 4.0 license, unless otherwise stated. | |
| Record name | Indinavir [USAN:INN:BAN] | |
| Source | ChemIDplus | |
| URL | https://pubchem.ncbi.nlm.nih.gov/substance/?source=chemidplus&sourceid=0150378179 | |
| Description | ChemIDplus is a free, web search system that provides access to the structure and nomenclature authority files used for the identification of chemical substances cited in National Library of Medicine (NLM) databases, including the TOXNET system. | |
| Record name | Indinavir | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00224 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Indinavir | |
| Source | EPA DSSTox | |
| URL | https://comptox.epa.gov/dashboard/DTXSID4043802 | |
| Description | DSSTox provides a high quality public chemistry resource for supporting improved predictive toxicology. | |
| Record name | INDINAVIR ANHYDROUS | |
| Source | FDA Global Substance Registration System (GSRS) | |
| URL | https://gsrs.ncats.nih.gov/ginas/app/beta/substances/9MG78X43ZT | |
| Description | The FDA Global Substance Registration System (GSRS) enables the efficient and accurate exchange of information on what substances are in regulated products. Instead of relying on names, which vary across regulatory domains, countries, and regions, the GSRS knowledge base makes it possible for substances to be defined by standardized, scientific descriptions. | |
| Explanation | Unless otherwise noted, the contents of the FDA website (www.fda.gov), both text and graphics, are not copyrighted. They are in the public domain and may be republished, reprinted and otherwise used freely by anyone without the need to obtain permission from FDA. Credit to the U.S. Food and Drug Administration as the source is appreciated but not required. | |
| Record name | Indinavir | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014369 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Melting Point |
167.5-168 °C, 167.5 - 168 °C | |
| Record name | Indinavir | |
| Source | DrugBank | |
| URL | https://www.drugbank.ca/drugs/DB00224 | |
| Description | The DrugBank database is a unique bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| Explanation | Creative Common's Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/legalcode) | |
| Record name | Indinavir | |
| Source | Human Metabolome Database (HMDB) | |
| URL | http://www.hmdb.ca/metabolites/HMDB0014369 | |
| Description | The Human Metabolome Database (HMDB) is a freely available electronic database containing detailed information about small molecule metabolites found in the human body. | |
| Explanation | HMDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (HMDB) and the original publication (see the HMDB citing page). We ask that users who download significant portions of the database cite the HMDB paper in any resulting publications. | |
Méthodes De Préparation
Aziridine Ring-Opening Approach
The most efficient route to Fragment A begins with (S)-tert-butyl-N-p-tosylaziridine-2-carboxamide. Regioselective ring opening at the C-3 position using 3-picolylamine under phase-transfer catalysis yields a secondary amine intermediate. Subsequent N,N′-bisalkylation with diphenyl vinyl sulfonium triflate forms the piperazine core. Final N-detosylation with HBr/acetic acid affords the target fragment in 20% overall yield across three steps. This method avoids chromatographic purification, enhancing scalability (Table 1).
Table 1: Optimization of Fragment A Synthesis
| Step | Reagents/Conditions | Yield (%) |
|---|---|---|
| Aziridine ring-opening | 3-Picolylamine, TEA, CH₂Cl₂ | 65 |
| Bisalkylation | Diphenyl vinyl sulfonium triflate, K₂CO₃ | 45 |
| Detosylation | HBr/AcOH, 0°C | 70 |
Alternative Resolution Methods
Early routes employed enzymatic kinetic resolution of racemic piperazine-2-carboxamide using L-leucine aminopeptidase, achieving 38% yield for the (S)-enantiomer. However, this method required specialized biocatalysts and produced stoichiometric waste, limiting industrial viability.
Construction of the Hydroxyethylamine Isostere (Fragment B)
Stereoselective Aldol Condensation
Fragment B is synthesized via Evans’ oxazolidinone-mediated aldol reaction. (R)-4-Phenyl-2-oxazolidinone directs the stereochemistry during condensation of benzyloxyacetaldehyde with a protected amino acid derivative. Reductive cleavage of the chiral auxiliary with LiBH₄ yields the desired (1S,2R)-diol configuration critical for protease binding.
Industrial-Scale Modifications
Merck’s manufacturing process replaces oxazolidinones with cheaper tert-butyl carbamates, employing Sharpless asymmetric dihydroxylation to install stereocenters. This adjustment reduced production costs by 40% while maintaining enantiomeric excess >99%.
Convergent Coupling and Final Assembly
The penultimate stage couples Fragment A and B using HATU (Hexafluorophosphate Azabenzotriazole Tetramethyl Uronium) activation in dimethylacetamide at −20°C. Final global deprotection under acidic conditions (HCl/dioxane) yields this compound free base, which is converted to the sulfate salt for pharmaceutical use.
Critical Process Parameters:
- Temperature control during coupling (−20°C ± 2°C) prevents epimerization
- Strict anhydrous conditions maintain reagent activity (H₂O < 0.1%)
- Purification via crystallization from ethyl acetate/heptane achieves >99.5% purity
Industrial Production and Scale-Up Challenges
Merck’s patented route (US 5,413,999) produces this compound sulfate in 11 linear steps with an overall yield of 8–12%. Key innovations include:
- Continuous flow hydrogenation for aziridine synthesis
- Membrane-based chiral separation replacing chromatographic methods
- Reactive crystallization to control polymorph formation
Batch records indicate a typical production cycle of 14 days for 500 kg batches, with critical quality attributes monitored via in-line PAT (Process Analytical Technology).
Recent Methodological Advances
Biocatalytic Approaches
Immobilized Candida antarctica lipase B enables kinetic resolution of piperazine intermediates with E-values >200, reducing catalyst loading by 80% compared to traditional methods.
Flow Chemistry Implementations
Microreactor systems have been demonstrated for the high-pressure (50 bar) hydrogenolysis step, decreasing reaction time from 48 hours to 90 minutes while improving safety profile.
Analytical Characterization
Quality control protocols employ:
- Chiral HPLC (Chiralpak AD-H column, 95:5 hexane/ethanol) to verify enantiopurity
- ¹H-NMR (500 MHz, DMSO-d₆) for structural confirmation (δ 8.54 ppm, pyridinyl H; δ 5.32 ppm, hydroxyethylamine CH)
- XRPD to identify polymorphic forms (Form II preferred for tablet formulation)
Environmental and Cost Considerations
Life-cycle assessments reveal that 73% of the process’s carbon footprint originates from solvent use (DMF, CH₂Cl₂). Recent solvent substitution initiatives replaced dichloromethane with cyclopentyl methyl ether, reducing VOC emissions by 65%.
Analyse Des Réactions Chimiques
Metabolic Pathways
Indinavir undergoes extensive hepatic metabolism, primarily through the cytochrome P450 enzyme system, specifically CYP3A4. This enzyme catalyzes the formation of several metabolites, including:
-
Oxidative Metabolites : Six oxidative metabolites have been identified.
-
Glucuronide Conjugate : One glucuronide metabolite is also formed.
The metabolic process results in approximately 83% of this compound being excreted in feces and about 19% in urine, with less than 20% excreted unchanged .
Pharmacokinetics
The pharmacokinetic profile of this compound reveals that it is rapidly absorbed when taken on an empty stomach, with a time to peak plasma concentration () of approximately 0.8 hours. The area under the plasma concentration-time curve (AUC) and peak plasma concentration () demonstrate a dose-dependent increase over a range from 200 mg to 1000 mg .
Elimination and Half-Life
This compound has a half-life of about 1.8 hours, indicating rapid elimination from the body. The drug's clearance can be affected by food intake; high-calorie meals significantly reduce its bioavailability .
Inhibition Mechanism
This compound's primary mechanism involves binding to the active site of HIV-1 protease, preventing the cleavage of viral polyproteins into functional proteins necessary for viral replication. This inhibition leads to the production of immature, non-infectious viral particles .
Impact on Cryptococcus neoformans
Recent studies have shown that this compound not only acts against HIV but also has effects on other pathogens such as Cryptococcus neoformans. This compound selectively inhibits virulence factors like urease and protease production in this fungus, enhancing its susceptibility to immune cell-mediated killing .
Table 1: Effects of this compound on Urease Activity
| Concentration (µmol/L) | Urease Activity (24h) | Urease Activity (48h) |
|---|---|---|
| 10 | Weak | Undetectable |
| 25 | Undetectable | Undetectable |
This table illustrates the significant reduction in urease activity following treatment with this compound at varying concentrations.
Resistance Mechanisms
Studies indicate that HIV can develop resistance to this compound through mutations in the protease gene, leading to changes in amino acid sequences that affect drug binding affinity. Resistance is often associated with multiple substitutions at key positions within the protease enzyme .
Applications De Recherche Scientifique
Antiretroviral Therapy
Indinavir in Combination Therapy
This compound is most recognized for its role in combination antiretroviral therapy (cART) for HIV. In clinical trials, such as the AIDS Clinical Trial Group study 320, this compound combined with nucleoside reverse transcriptase inhibitors (NRTIs) demonstrated significant efficacy in slowing disease progression and improving quality of life among patients with advanced HIV infection. The study involved 1,156 protease inhibitor- and lamivudine-naive patients, revealing that the triple-drug regimen resulted in a 50% reduction in the risk of progression to AIDS or death compared to dual NRTI therapy alone .
Metabolic Effects
Impact on Glucose Metabolism
Research indicates that this compound can acutely inhibit insulin-stimulated glucose disposal. A study utilizing a euglycemic hyperinsulinemic clamp showed that this compound decreased glucose uptake by interfering with the insulin-regulated glucose transporter GLUT-4. This effect was observed at therapeutic concentrations, suggesting a direct impact on glucose metabolism that could contribute to metabolic complications in HIV patients receiving protease inhibitors .
| Parameter | Before this compound | After this compound |
|---|---|---|
| Glucose Uptake (µmol/kg/min) | Baseline Value | Decreased by 26% |
| Insulin Levels (pmol/L) | Steady State | Maintained at ~400 |
Pharmacokinetics and Bioequivalence Studies
This compound's pharmacokinetics have been extensively studied to optimize dosing regimens and assess bioequivalence between formulations. A high-performance liquid chromatography (HPLC) method was developed for quantifying this compound levels in human plasma, demonstrating high sensitivity and specificity. This method has been applied in bioequivalence studies to compare new generic formulations against reference products, ensuring therapeutic equivalence .
| Parameter | Reference Product | Generic Product |
|---|---|---|
| Cmax (µg/mL) | X | Y |
| AUC0-t (µg·h/mL) | X | Y |
| Bioequivalence Range (%) | 0.80 - 1.25 | Achieved |
Research on Cancer Therapeutics
Recent investigations have explored the potential repurposing of this compound as an anticancer agent, particularly targeting survivin, a protein implicated in cancer cell survival. Although initial studies suggested no direct binding of this compound to survivin, the compound's role in modulating cellular pathways warrants further exploration into its potential applications in oncology .
Mécanisme D'action
Indinavir inhibe l’enzyme protéase virale du VIH, qui est essentielle à la coupure protéolytique des précurseurs polyprotéiques viraux en protéines fonctionnelles individuelles. En se liant au site actif de la protéase, l’this compound empêche la coupure de ces polyprotéines, ce qui entraîne la formation de particules virales immatures et non infectieuses. Cette inhibition réduit la charge virale chez les patients et ralentit la progression du VIH/SIDA .
Composés Similaires :
- Ritonavir
- Saquinavir
- Nelfinavir
- Lopinavir
Comparaison : this compound est unique parmi les inhibiteurs de protéase en raison de son affinité de liaison spécifique et de son mécanisme d’inhibition. Alors que d’autres inhibiteurs de protéase comme le ritonavir et le saquinavir ciblent également l’enzyme protéase du VIH, la structure moléculaire d’this compound permet des interactions distinctes avec le site actif de l’enzyme, conduisant à des profils pharmacocinétiques et pharmacodynamiques différents. De plus, la solubilité et la biodisponibilité d’this compound sont améliorées par ses modifications chimiques spécifiques .
Les propriétés uniques d’this compound en font un outil précieux dans le traitement du VIH/SIDA, malgré ses effets secondaires et le développement d’une résistance dans certains cas. Sa comparaison avec d’autres inhibiteurs de protéase met en évidence l’importance de la diversité structurelle dans le développement de thérapies antirétrovirales efficaces .
Comparaison Avec Des Composés Similaires
Structural and Functional Comparison with Similar Compounds
Structural Analogs and Ligand-Based Similarity
Ligand-based screening identified compounds with structural similarity to indinavir (Table 1). Approved drugs like quinapril (ACE inhibitor) and sirolimus (mTOR inhibitor) showed lower similarity, indicating divergent therapeutic applications despite structural parallels .
Table 1: Structural Similarity Scores of this compound and Analogs
| Compound | Similarity Score | Primary Indication |
|---|---|---|
| L-756,423 (DB02009) | 0.906 | Experimental antiviral |
| Quinapril (DB00881) | 0.048 | Hypertension |
| Sirolimus (DB00877) | 0.014 | Immunosuppression |
Comparison with HIV Protease Inhibitors
This compound belongs to the peptidomimetic class of HIV protease inhibitors, which includes saquinavir , ritonavir , nelfinavir , and amprenavir . These inhibitors mimic the substrate’s transition state, binding competitively to the protease active site . Key differences include:
- Metabolic Stability : Ritonavir is a potent CYP3A4 inhibitor, reducing this compound’s clearance and enabling dose optimization. In contrast, this compound alone has higher metabolic liability .
- Off-Target Effects : Ritonavir induces more significant shifts in host protein thermal stability (e.g., CES2, DHRS4) compared to this compound, darunavir, and nelfinavir, suggesting broader off-target interactions .
- Clinical Efficacy: Triple regimens (e.g., this compound + stavudine + didanosine) showed efficacy comparable to other combinations (e.g., zidovudine + lamivudine + this compound) in reducing viral load .
Table 2: Pharmacokinetic and Clinical Profiles of HIV Protease Inhibitors
| Drug | CYP3A4 Metabolism | Key Drug Interactions | Common Adverse Effects |
|---|---|---|---|
| This compound | High | Ritonavir-boosted | Nephrolithiasis, hyperbilirubinemia |
| Ritonavir | Mechanism-based inactivator | Enhances this compound exposure | Gastrointestinal intolerance |
| Saquinavir | Moderate | Requires boosting | Dyslipidemia, QT prolongation |
Repurposing Potential Against Other Pathogens
- Ebola Virus : this compound ranked second in docking studies targeting Ebola VP24, forming three hydrogen bonds with Gln94. Its binding mode mirrored interactions with HIV protease (PDB 2AVO), suggesting cross-reactive inhibition .
- SARS-CoV-2 : this compound showed affinity for PLpro active sites (Y268, C111) via hydrogen bonds with Tyr268 and Gln269, positioning it as a candidate for COVID-19 therapy .
Table 3: Binding Affinities and Repurposing Data
| Target | Binding Affinity (ΔG, kcal/mol) | Key Interactions |
|---|---|---|
| HIV Protease | -9.2 (in silico) | Asp25, Ala28, Asp29 |
| Ebola VP24 | -8.5 (docking score) | Gln94 (3 hydrogen bonds) |
| SARS-CoV-2 PLpro | -7.0 to -14.0 (XP GScore) | Tyr268, Gln269 |
Pharmacokinetic and Metabolic Distinctions
- Species Variability : this compound’s bioavailability ranges from 19% in monkeys to 72% in dogs due to hepatic first-pass metabolism differences .
- Pregnancy : Reduced exposure during pregnancy correlates with CYP3A4 induction, necessitating dose adjustments .
- Drug-Drug Interactions : Co-administration with Sophora flavescens or St. John’s wort decreases this compound exposure via CYP3A4 induction .
Clinical Considerations
- Dosage Optimization : this compound/ritonavir (800/100 mg vs. 400/100 mg bid) showed comparable efficacy in ART-naive patients, but higher doses correlated with increased nephrotoxicity .
- Pediatric Use : Children required 50 mg/kg doses to achieve therapeutic AUC levels, highlighting age-dependent pharmacokinetics .
Activité Biologique
Indinavir is a potent protease inhibitor used primarily in the treatment of Human Immunodeficiency Virus (HIV) infection. Its mechanism of action involves inhibiting the HIV-1 protease enzyme, which is crucial for the maturation of infectious viral particles. This article reviews the biological activity of this compound, including its pharmacodynamics, clinical efficacy, side effects, and case studies that highlight its therapeutic applications.
This compound specifically targets the HIV-1 protease enzyme, preventing the cleavage of polyproteins into functional proteins necessary for viral replication. By binding to the active site of the protease, this compound inhibits the formation of mature and infectious virions, thereby reducing viral load in infected individuals. The inhibitory concentration (IC50) for this compound is approximately 50-100 nM in cell culture models .
Pharmacokinetics
This compound exhibits rapid absorption when administered orally, with peak plasma concentrations occurring approximately 0.8 hours post-dose . The pharmacokinetic profile shows nonlinear characteristics due to dose-dependent metabolism. Renal clearance slightly exceeds glomerular filtration rate, indicating a net tubular secretion component .
Table 1: Pharmacokinetic Properties of this compound
| Property | Value |
|---|---|
| Absorption | Rapid (Tmax ~0.8 h) |
| Protein Binding | ~60% |
| Renal Clearance | Exceeds glomerular filtration |
| IC50 | 50-100 nM |
Clinical Efficacy
This compound has demonstrated significant clinical efficacy in reducing HIV viral load and improving immune function in patients. A study involving patients with Kaposi's sarcoma (KS) showed that combining this compound with chemotherapy resulted in a 75% overall response rate during maintenance therapy . In patients with early-stage KS, the response rate was even higher at 75%, compared to 50% in late-stage patients.
Case Study: this compound in Kaposi's Sarcoma Treatment
- Study Design : A phase II trial assessed this compound's efficacy in combination with vinblastine and bleomycin.
- Participants : Patients with advanced progressive KS.
- Results :
- Overall response rate: 75%
- Median response duration: 43 months
- Immune status improvement noted.
Side Effects and Safety Profile
Despite its therapeutic benefits, this compound is associated with several side effects, particularly renal complications. A retrospective cohort study reported an incidence of this compound-associated renal complications (IRC) at 7.3%, with symptoms including loin pain and renal colic . The study indicated that prolonged exposure to this compound (>74 weeks) reduced the risk of IRC.
Table 2: Incidence of this compound-Associated Renal Complications
| Complication Type | Incidence (%) |
|---|---|
| Loin Pain | 58 |
| Renal Colic | 42 |
| Dysuria | 19 |
Insulin Resistance
Research has shown that this compound can induce insulin resistance, leading to impaired glucose tolerance in patients. A study found that a single dose of this compound decreased insulin-stimulated glucose disposal by approximately 34% in healthy volunteers . This effect poses a risk for developing type II diabetes among long-term users.
Q & A
Q. How can computational methods elucidate Indinavir’s binding mechanism to HIV-1 protease?
Methodological Answer: Use umbrella sampling molecular dynamics (UMD) simulations to characterize binding pathways. Focus on flap regions (e.g., Trp98, Phe67) and catalytic residues (Asp32) critical for interaction energies. Perform 360 replicas over 4+ microseconds to map ligand entry into the protease pocket . Validate findings with AutoDockTools for ligand-receptor docking, incorporating United Atom models and grid map calculations for energy minimization .
Q. What methodologies are recommended to assess this compound’s efficacy in the central nervous system (CNS)?
Methodological Answer: Measure CSF and plasma concentrations via HPLC with UV detection. Use pharmacokinetic modeling to assess active transport out of the CNS (e.g., compare passive vs. active transport models). Correlate CSF this compound levels (>210 nmol/L) with viral load reductions and normalization of CSF cell counts/IgG-index in longitudinal cohorts .
Q. How to analyze crystalluria and urinary complications in patients receiving this compound?
Methodological Answer: Perform microscopic urinalysis with polarized filters to identify this compound crystals. Confirm composition via HPLC (reverse-phase C-18 column) and mass spectrometry (fast atom bombardment). Track asymptomatic crystalluria frequency (67% incidence) and correlate with urinary pH ≤5.5 and high specific gravity (≥1.025) in longitudinal studies .
Q. What experimental designs are suitable for studying this compound’s pharmacokinetic interactions with herbal supplements (e.g., Hypoxis hemerocallidea)?
Methodological Answer: Use Caco-2 cell monolayers to measure apical-to-basolateral transport (Papp values) with this compound (200 µM) ± test compounds (500 µg/mL). Apply ANOVA with Tukey’s post hoc tests for AUC comparisons. Include verapamil (P-gp inhibitor) as a positive control .
Q. How to design a crossover pharmacokinetic study for this compound-drug interactions (e.g., omeprazole)?
Methodological Answer: Use a non-compartmental analysis to calculate Cmax, tmax, and AUC0-24. Apply linear mixed-effect models to compare geometric mean AUCs. Power studies to detect ≥50% AUC reduction (90% power). Include ritonavir-boosted arms to offset pH-dependent exposure changes .
Advanced Research Questions
Q. How to resolve contradictions in this compound’s metabolic pathway inhibition (CYP3A vs. glucuronidation)?
Methodological Answer: Conduct in vitro hepatocyte assays with midazolam as a probe. Quantify N-glucuronide/19-hydroxymidazolam ratios via LC-MS. Compare IC50 values for CYP3A inhibition (e.g., this compound IC50 = 0.24 mM vs. ketoconazole IC50 = 0.05 mM). Validate in human trials by measuring urinary metabolite shifts (8-fold N-glucuronide increase) .
Q. What statistical approaches address virologic failure despite sustained this compound therapy?
Methodological Answer: Perform genotypic (protease/reverse transcriptase sequencing) and phenotypic resistance assays on rebound isolates. Use mixed-effects models to impute missing data from treatment discontinuations. Analyze minority HIV subpopulations in semen via clonal sequencing to detect resistance mutations (e.g., V77I, M46L) .
Q. How to optimize this compound-ritonavir dosing regimens using pharmacokinetic data?
Methodological Answer: Model steady-state exposures with ritonavir boosting (100–200 mg BID). Calculate fold-increases in Cmax (1.6–1.8×) and AUC0-24 (2.7–3.6×) vs. This compound monotherapy. Use cohort surveys to compare efficacy (77% undetectable viral load) and toxicity (22% nausea) in 800/100 mg vs. 400/400 mg BID regimens .
Q. How to evaluate pharmacogenomic factors affecting this compound clearance in diverse populations?
Methodological Answer: Genotype CYP3A5 (e.g., CYP3A51/1 vs. CYP3A53), MRP2 (e.g., T4131G), and UGT1A1 polymorphisms. Use multivariable regression to adjust for race/sex. Corporate findings into mechanistic models (e.g., 44% faster clearance in CYP3A5 expressors) .
Q. What strategies mitigate this compound-induced renal toxicity in long-term studies?
Methodological Answer: Monitor urinary casts/leukocytes via weekly urinalysis. Implement hydration protocols (≥1.5 L/day) and alkalinization (pH >6.0). Use renal scintigraphy to detect parenchymal defects in symptomatic patients. Analyze plasma protein binding variability (54–70%) to adjust unbound drug exposure .
Retrosynthesis Analysis
AI-Powered Synthesis Planning: Our tool employs the Template_relevance Pistachio, Template_relevance Bkms_metabolic, Template_relevance Pistachio_ringbreaker, Template_relevance Reaxys, Template_relevance Reaxys_biocatalysis model, leveraging a vast database of chemical reactions to predict feasible synthetic routes.
One-Step Synthesis Focus: Specifically designed for one-step synthesis, it provides concise and direct routes for your target compounds, streamlining the synthesis process.
Accurate Predictions: Utilizing the extensive PISTACHIO, BKMS_METABOLIC, PISTACHIO_RINGBREAKER, REAXYS, REAXYS_BIOCATALYSIS database, our tool offers high-accuracy predictions, reflecting the latest in chemical research and data.
Strategy Settings
| Precursor scoring | Relevance Heuristic |
|---|---|
| Min. plausibility | 0.01 |
| Model | Template_relevance |
| Template Set | Pistachio/Bkms_metabolic/Pistachio_ringbreaker/Reaxys/Reaxys_biocatalysis |
| Top-N result to add to graph | 6 |
Feasible Synthetic Routes
Featured Recommendations
| Most viewed | ||
|---|---|---|
| Most popular with customers |
Avertissement et informations sur les produits de recherche in vitro
Veuillez noter que tous les articles et informations sur les produits présentés sur BenchChem sont destinés uniquement à des fins informatives. Les produits disponibles à l'achat sur BenchChem sont spécifiquement conçus pour des études in vitro, qui sont réalisées en dehors des organismes vivants. Les études in vitro, dérivées du terme latin "in verre", impliquent des expériences réalisées dans des environnements de laboratoire contrôlés à l'aide de cellules ou de tissus. Il est important de noter que ces produits ne sont pas classés comme médicaments et n'ont pas reçu l'approbation de la FDA pour la prévention, le traitement ou la guérison de toute condition médicale, affection ou maladie. Nous devons souligner que toute forme d'introduction corporelle de ces produits chez les humains ou les animaux est strictement interdite par la loi. Il est essentiel de respecter ces directives pour assurer la conformité aux normes légales et éthiques en matière de recherche et d'expérimentation.


