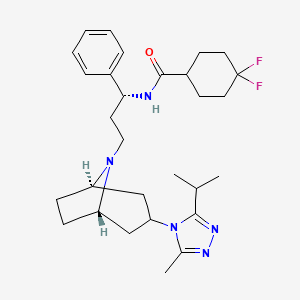
Maraviroc
Description
Historical Context of HIV-1 Entry Inhibition Research
Research into blocking HIV-1 entry began with exploring molecules that could interfere with the interaction between the viral envelope protein gp120 and the host cell's primary receptor, CD4. Early attempts included the development of recombinant soluble CD4 molecules, designed to act as decoys for gp120. nih.gov While these showed in vitro activity against certain lab-adapted HIV-1 strains, their efficacy in clinical trials was limited, partly because primary HIV-1 isolates were less sensitive to neutralization by these molecules. nih.gov The subsequent identification of chemokine receptors as essential co-receptors for HIV-1 entry opened new avenues for therapeutic intervention. nih.govbioline.org.br
Role of Chemokine Receptors in HIV-1 Tropism and Entry
HIV-1 entry into target cells is a complex process that requires the sequential interaction of the viral envelope glycoprotein (Env), specifically the gp120 subunit, with the primary receptor CD4 and a chemokine receptor, which acts as a co-receptor. nih.govembopress.org The choice of co-receptor is a major determinant of viral tropism, influencing which cell types the virus can efficiently infect. nih.govnih.gov
The C-C chemokine receptor type 5 (CCR5) was identified as a major co-receptor for primary HIV-1 isolates. scientificarchives.compnas.org CCR5 is a G protein-coupled receptor expressed on various immune cells, including activated T lymphocytes, macrophages, and dendritic cells. scientificarchives.com Interaction with CCR5 is critical for the entry of macrophage-tropic (M-tropic) or R5 strains of HIV-1, which are predominantly responsible for viral transmission and the establishment of initial infection in humans. pnas.orgaai.orgnih.gov The importance of CCR5 is underscored by the observation that individuals homozygous for a natural loss-of-function mutation in the CCR5 gene, known as CCR5-Δ32, are highly resistant to HIV-1 infection. scientificarchives.comaai.orgnih.gov This genetic evidence provided strong support for targeting CCR5 to inhibit viral entry. scientificarchives.com
In addition to CCR5, the C-X-C chemokine receptor type 4 (CXCR4) also serves as a co-receptor for HIV-1 entry. pnas.orgaai.org Viruses that utilize CXCR4 are referred to as T-cell tropic (T-tropic) or X4 strains. pnas.org Unlike R5 strains which dominate early infection, X4 strains often emerge later during the course of disease progression, particularly in individuals with advanced HIV-1 infection and lower CD4+ T cell counts. aai.orgtandfonline.comdaig-net.de The emergence of X4 viruses is frequently associated with accelerated immunologic decline and faster progression to AIDS. tandfonline.comasm.orgnih.gov Some HIV-1 strains can utilize both CCR5 and CXCR4 for entry; these are termed dual-tropic or mixed-tropic viruses. pnas.orgasm.orgmdpi.com The presence of dual/mixed-tropic populations or CXCR4-using virus can occur across a range of CD4 cell counts and viral loads but is more common with lower CD4 counts and higher viral loads. tandfonline.com
CCR5 as a Primary Co-receptor for HIV-1 Entry
Maraviroc as a First-in-Class CCR5 Antagonist
Building on the understanding of CCR5's crucial role in HIV-1 entry, research efforts were directed towards developing small molecules that could block this interaction. This compound (originally known as UK-427,857) emerged from a high-throughput screening program designed to identify compounds that could prevent the binding of chemokine ligands to the CCR5 receptor. frontiersin.orgnih.gov
This compound is a selective, orally bioavailable small-molecule CCR5 antagonist. asm.org Its mechanism of action involves binding to the transmembrane pocket of CCR5, inducing an allosteric modification that disrupts the necessary interaction between the viral gp120 protein and the CCR5 co-receptor. nih.govasm.org This binding prevents the conformational changes in the viral envelope proteins required for the fusion of the viral membrane with the host cell membrane, thereby blocking viral entry. nih.govasm.orgdrugbank.com
This compound was the first CCR5 antagonist to receive approval for the treatment of HIV-1 infection. scientificarchives.comoup.comnih.gov It demonstrated potent antiviral activity against a broad range of CCR5-tropic HIV-1 isolates in in vitro studies. asm.org Research findings indicated that this compound effectively inhibited the entry of R5 strains into target cells. nih.gov Its development marked a significant milestone, representing a new class of antiretroviral agents that target a host protein rather than a viral enzyme. nih.govoup.com
Interactive Table: HIV-1 Coreceptors and Tropism
| Co-receptor | Associated Tropism | Prevalence in Infection | Disease Progression Association |
| CCR5 | R5 (Macrophage-tropic) | Predominant in early infection | Generally slower progression |
| CXCR4 | X4 (T-cell tropic) | Emerges later in ~50% of clade B infections | Associated with accelerated progression |
| Both (CCR5 and CXCR4) | Dual/Mixed-tropic | Can be present at various stages, more common in advanced disease | Associated with accelerated progression |
This table is for illustrative purposes based on the text and can be made interactive in a suitable digital format.
Properties
IUPAC Name |
4,4-difluoro-N-[3-[3-(3-methyl-5-propan-2-yl-1,2,4-triazol-4-yl)-8-azabicyclo[3.2.1]octan-8-yl]-1-phenylpropyl]cyclohexane-1-carboxamide | |
|---|---|---|
| Details | Computed by Lexichem TK 2.7.0 (PubChem release 2021.05.07) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI |
InChI=1S/C29H41F2N5O/c1-19(2)27-34-33-20(3)36(27)25-17-23-9-10-24(18-25)35(23)16-13-26(21-7-5-4-6-8-21)32-28(37)22-11-14-29(30,31)15-12-22/h4-8,19,22-26H,9-18H2,1-3H3,(H,32,37) | |
| Details | Computed by InChI 1.0.6 (PubChem release 2021.05.07) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
InChI Key |
GSNHKUDZZFZSJB-UHFFFAOYSA-N | |
| Details | Computed by InChI 1.0.6 (PubChem release 2021.05.07) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Canonical SMILES |
CC1=NN=C(N1C2CC3CCC(C2)N3CCC(C4=CC=CC=C4)NC(=O)C5CCC(CC5)(F)F)C(C)C | |
| Details | Computed by OEChem 2.3.0 (PubChem release 2021.05.07) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Formula |
C29H41F2N5O | |
| Details | Computed by PubChem 2.1 (PubChem release 2021.05.07) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Molecular Weight |
513.7 g/mol | |
| Details | Computed by PubChem 2.1 (PubChem release 2021.05.07) | |
| Source | PubChem | |
| URL | https://pubchem.ncbi.nlm.nih.gov | |
| Description | Data deposited in or computed by PubChem | |
Discovery and Preclinical Development of Maraviroc
High-Throughput Screening Initiatives for CCR5 Ligands
High-throughput screening played a crucial role in the initial identification of potential CCR5 ligands nih.govtandfonline.com. Pfizer screened its compound library, which contained approximately 500,000 molecules, using a chemokine radioligand-binding assay nih.govtandfonline.com. This assay utilized HEK-293 cells expressing CCR5 and measured the inhibition of radiolabeled MIP-1β binding nih.govtandfonline.com.
Identification of Initial Hit Compounds
The initial HTS efforts identified a number of hit compounds nih.gov. These hits were then triaged based on factors such as target affinity, ligand efficiency, and toxicity nih.gov. After this initial selection process, four hits remained nih.gov. Two of these, imidazopyridine 1 (UK-107,543) and 2 (UK-179,645), were chosen as starting points for further intensive medicinal chemistry optimization nih.gov. Compound 1 showed a MIP-1β IC₅₀ of 0.4 μM, while compound 2 had a MIP-1β IC₅₀ of 1.1 μM nih.gov. However, these initial hits lacked desirable properties such as low molecular weight, high binding affinity, and potent antiviral activity, necessitating further optimization nih.gov.
Hit-to-Lead Optimization Strategies
The hit-to-lead optimization phase aimed to improve the properties of the initial hit compounds nih.gov. This involved combining the attractive features of the two initial hits to generate novel and selective antagonists nih.gov. Compound 3 emerged as a key focus for structure-activity relationship (SAR) investigations during this stage nih.gov. Compound 3 demonstrated improved chemokine receptor binding with a MIP-1β IC₅₀ of 45 nM and exhibited antiviral activity nih.gov. Lead optimization efforts also focused on addressing potential issues such as hERG liability nih.gov.
Medicinal Chemistry Efforts and Analog Synthesis
Extensive medicinal chemistry efforts were undertaken in the development of Maraviroc, involving the synthesis of nearly 1,000 analogues nih.gov. This iterative process aimed to optimize the pharmacological and pharmacokinetic properties of the identified chemical series asm.org.
Structure-Activity Relationship (SAR) Investigations
Structure-activity relationship (SAR) investigations were central to the optimization process nih.govscirp.org. These studies explored the relationship between the chemical structure of the synthesized analogues and their biological activity, including receptor binding and antiviral potency nih.govscirp.org. SAR analysis helped guide modifications to the lead compounds to enhance their efficacy and selectivity nih.gov.
Elucidation of Key Pharmacophore Elements
SAR studies provided insights into the key pharmacophore elements necessary for effective CCR5 binding and antagonism nih.govacs.org. For instance, SAR suggested that the amide substituent in the lead series interacts with a predominantly lipophilic binding site within the CCR5 receptor nih.gov. The crystal structure of CCR5 bound to this compound, published in 2013, provided a structural basis for understanding the binding site, revealing an allosteric binding pocket located at the extracellular end of the transmembrane bundle scirp.orgacs.orgnih.gov. This pocket occupies both the transmembrane site 1 (minor pocket) and transmembrane site 2 (major pocket) acs.org.
Optimization for Receptor Binding and Antiviral Potency
Optimization efforts focused on improving both the binding affinity to the CCR5 receptor and the antiviral potency against HIV-1 nih.gov. Modifications were made to the lead compounds based on SAR data to enhance these properties nih.gov. For example, the cyclobutyl amide analogue 4 was identified as the most potent in its series based on SAR studies of different amide substituents nih.gov. The final compound, this compound (originally known as UK-427,857), was identified after approximately two and a half years of development and the synthesis of 965 analogues nih.govnih.gov. It demonstrated potent antiviral activity against CCR5-tropic HIV-1 viruses, with a geometric mean 90% inhibitory concentration (IC90) of 2.0 nM against a diverse panel of primary isolates asm.orgresearchgate.net. This compound selectively binds to CCR5, preventing the interaction of the viral envelope protein gp120 with the receptor, which is necessary for viral entry asm.org. This mechanism of action was confirmed through cell-based assays asm.org.
Data Table: Initial Hit Compounds and a Lead Compound
| Compound | Identification | MIP-1β IC₅₀ (μM) | Antiviral Activity |
| UK-107,543 (1) | HTS Hit | 0.4 | Not desirable |
| UK-179,645 (2) | HTS Hit | 1.1 | Not desirable |
| Compound 3 | Lead Compound | 0.045 (45 nM) | Good |
Note: IC₅₀ values indicate the concentration required for 50% inhibition of MIP-1β binding. nih.gov
Lead Optimization for Selectivity and Pharmacological Properties
Following the identification of initial lead compounds, an intensive medicinal chemistry program was initiated to optimize their properties. asm.org The primary goals of this lead optimization process were to enhance binding potency against the CCR5 receptor, improve antiviral activity, and optimize absorption and pharmacokinetic profiles. asm.org Crucially, significant effort was directed towards ensuring selectivity against key human targets, particularly the hERG ion channel, to mitigate potential cardiac liabilities. asm.orgdovepress.comnih.gov
The optimization process involved the synthesis and profiling of nearly 1,000 analogues. asm.org Parallel screening was employed to evaluate compounds based on multiple parameters simultaneously. asm.org Bespoke assays were developed and optimized to measure envelope binding to cell surface receptors and model subsequent membrane fusion events, mimicking HIV-1 entry into host cells. asm.org
Challenges addressed during lead optimization included improving the binding efficiency of screening hits, reducing inhibition of CYP2D6, and minimizing potent inhibition of the hERG cardiac potassium channel. dovepress.com The development of a new class of CCR5 antagonist, replacing the tropane core of an earlier series with a piperidine with a branched N-substituent, contributed to identifying compounds with good whole-cell antiviral activity, excellent selectivity over hERG, and complete oral absorption. researchgate.net this compound was ultimately selected from this extensive series of analogues due to its desirable antiviral activity, metabolic stability, absorption characteristics, and lack of significant hERG inhibition. asm.orgnih.gov
This compound demonstrated potent inhibition of MIP-1β binding to the CCR5 receptor with an IC50 of 2 nM and potent antiviral activity with an EC90 of 1 nM against HIVBAL replication in PM1 cells, while showing only modest inhibition of the hERG potassium channel. dovepress.com
Preclinical Evaluation and Nomination as a Clinical Candidate
Extensive preclinical evaluations were conducted to assess the pharmacological profile and potential of this compound as a clinical candidate. This compound was confirmed as a selective CCR5 antagonist with potent anti-HIV-1 activity and favorable pharmacological properties. asm.orgresearchgate.net
In cell-based assays, this compound was shown to block the binding of viral envelope protein gp120 to CCR5, thereby preventing the membrane fusion necessary for viral entry. asm.orgresearchgate.net It inhibited chemokine binding and CCR5-mediated signaling, as demonstrated in Ca2+ mobilization and γ-S-GTP binding assays. asm.org this compound did not affect CCR5 cell surface levels or associated intracellular signaling, confirming its function as a CCR5 antagonist. asm.orgresearchgate.net
This compound exhibited potent antiviral activity against a broad spectrum of CCR5-tropic HIV-1 viruses. asm.orgresearchgate.net This included 43 primary isolates from various clades and diverse geographic origins, with a geometric mean 90% inhibitory concentration (IC90) of 2.0 nM. asm.orgresearchgate.net The IC90 values ranged from 0.5 nM to 13.4 nM. asm.org this compound also demonstrated activity against 200 clinically derived HIV-1 envelope-recombinant pseudoviruses, including those resistant to existing drug classes, with little difference in sensitivity. asm.orgresearchgate.net
Preclinical in vitro studies indicated no detectable cytotoxicity for this compound and confirmed its high selectivity for CCR5 over a wide range of receptors and enzymes, including the hERG ion channel (50% inhibitory concentration, >10 μM). asm.orgresearchgate.net
Pharmacokinetic studies were conducted in preclinical in vivo models, such as rats and dogs, following single intravenous and oral administrations. asm.org These studies predicted that this compound would have human pharmacokinetics consistent with once- or twice-daily oral administration. asm.orgresearchgate.net Preclinical data also indicated low cerebrospinal fluid exposure in rats, approximately 10% of free plasma concentrations. tga.gov.au this compound was found to be principally metabolized by the cytochrome P450 system, with CYP3A4 identified as the major metabolizing enzyme. tga.gov.au
Based on its potent antiviral activity against CCR5-tropic HIV-1, favorable pharmacological properties, excellent preclinical safety profile, and acceptable pharmacokinetics, this compound was nominated as a clinical candidate in December 2000. nih.gov
Preclinical Antiviral Activity of this compound against Primary CCR5-tropic HIV-1 Isolates asm.org
| Parameter | Value | 95% Confidence Interval |
| Geometric Mean IC90 (nM) | 2.0 | 1.8 to 2.4 |
| Range of IC90 (nM) | 0.5 to 13.4 |
This compound Activity in CCR5 Binding and Functional Assays asm.org
| Assay Type | Ligand/Mediator | IC50 (nM) | 95% Confidence Interval |
| Radioligand Binding | MIP-1α | 3.3 | 1.9 to 5.7 |
| Radioligand Binding | MIP-1β | 7.2 | 5.5 to 9.5 |
| Radioligand Binding | RANTES | 5.2 | 2.1 to 13 |
| Cell-based fusion assay | HIV-1 gp120 | 0.22 | 0.13 to 0.39 |
Preclinical Pharmacokinetic Data (Representative) asm.org
| Species | Administration Route | Dose (mg/kg) |
| Rat | Intravenous | 1 |
| Rat | Oral | 10 |
| Dog | Intravenous | 0.5 |
| Dog | Oral | 2 |
Molecular Mechanisms of Maraviroc Action
Selective CCR5 Binding and Antagonism
Maraviroc selectively binds to the human chemokine receptor CCR5. drugbank.compmda.go.jpasm.org This binding is characterized as slowly reversible and selective. hivclinic.ca By binding to CCR5, this compound functions as an antagonist, inhibiting the interaction between the HIV-1 envelope glycoprotein gp120 and CCR5. wikipedia.orgdrugbank.compmda.go.jphivclinic.caasm.org This selective binding is crucial as it prevents a necessary step in the entry process for CCR5-tropic HIV-1 strains. pmda.go.jp Studies have confirmed the high affinity and specificity of this compound binding to human CCR5 through radioligand binding experiments. pmda.go.jp
This compound acts as a negative allosteric modulator of the CCR5 receptor. wikipedia.orgnih.govscirp.org Allosteric modulation involves binding to a site on the receptor distinct from the orthosteric binding site where natural ligands or the viral protein would typically bind. nih.govresearchgate.net This allosteric binding leads to changes in the receptor's conformation. patsnap.comresearchgate.netnih.govasm.orgnih.gov Unlike natural β-chemokine CCR5 ligands, which induce signaling and internalization of the receptor, this compound-bound CCR5 does not signal and remains on the cell surface. researchgate.net
Binding of this compound to CCR5 induces conformational changes in the receptor. patsnap.comresearchgate.netnih.govasm.orgnih.gov this compound is thought to bind within a hydrophobic cavity formed by the transmembrane helices of CCR5. researchgate.netasm.orgdovepress.com This interaction, involving amino acids such as Trp86, Glu283, Tyr108, Tyr251, and Ile198, leads to conformational changes, particularly in the extracellular loops of CCR5. researchgate.netnih.govnih.gov These drug-induced changes result in a receptor conformation that is not recognized by the HIV-1 gp120 protein. researchgate.netnih.gov Different CCR5 antagonists can induce distinct conformational changes in the extracellular loops, suggesting drug-specific alterations. asm.org
The conformational changes induced by this compound binding disrupt the interaction between the HIV-1 gp120 protein and CCR5. patsnap.comwikipedia.orgpmda.go.jpresearchgate.netoup.comnih.gov By altering the shape of CCR5, particularly its extracellular loops, this compound prevents the proper association of gp120 with the co-receptor. researchgate.netnih.govnih.gov This disruption is a key step in blocking viral entry. patsnap.com
This compound directly inhibits the binding of HIV-1 gp120 to CCR5. wikipedia.orgpmda.go.jpasm.orgresearchgate.netoup.comnih.gov This inhibition has been demonstrated in cell-based assays where this compound blocked the binding of viral envelope protein gp120 to CCR5-expressing cells. asm.org The ability of this compound to inhibit soluble gp120 binding to CCR5-expressing cells has been characterized, with studies determining the IC50 for this inhibition. asm.org
| Assay Type | Analyte/Interaction Inhibited | IC50 (nM) | 95% Confidence Interval (nM) | Reference |
| Radioligand binding competition assay | MIP-1α binding to CCR5 | 3.3 | 1.9 to 5.7 | asm.org |
| Radioligand binding competition assay | MIP-1β binding to CCR5 | 7.2 | 5.5 to 9.5 | asm.org |
| Radioligand binding competition assay | RANTES binding to CCR5 | 5.2 | 2.1 to 13 | asm.org |
| Inhibition of soluble gp120 binding to CCR5 | gp120-sCD4 complex binding | 11 | 10.4 to 11.6 | asm.org |
| Inhibition of HIV-1 envelope protein-CCR5-dependent cell-cell fusion | Cell-cell fusion | 0.22 | 0.13 to 0.39 | asm.org |
| Inhibition of gp120 binding to CCR5 | gp120 binding | 6.4 | - | tocris.com |
By inhibiting the binding of gp120 to CCR5, this compound effectively prevents the subsequent steps required for viral entry, including the fusion of the viral membrane with the host cell membrane. asm.orgresearchgate.netoup.comnih.govnih.govpixorize.comjst.go.jp The interaction between gp120 and CCR5, after initial binding to the CD4 receptor, triggers conformational changes in the viral protein gp41, which are necessary for membrane fusion and entry of the viral core into the target cell. pmda.go.jpresearchgate.net this compound blocks this process by interfering with the gp120-CCR5 interaction. pmda.go.jpasm.orgresearchgate.net
Induced Conformational Changes in CCR5
Inhibition of HIV-1 gp120 Binding to CCR5
Impact on Chemokine Binding and Signaling
This compound also impacts the binding of natural chemokines to CCR5 and subsequent intracellular signaling. asm.orgresearchgate.netresearchgate.netwikipedia.org CCR5 is a receptor for several endogenous chemokines, including MIP-1α (CCL3), MIP-1β (CCL4), and RANTES (CCL5). pmda.go.jpasm.orgnih.gov this compound acts as a selective antagonist of the binding and intracellular signaling mediated by these chemokines. pmda.go.jp
Studies have shown that this compound inhibits the binding of MIP-1α, MIP-1β, and RANTES to CCR5 in radioligand binding competition assays. asm.org This inhibition of chemokine binding also prevents downstream CCR5 signaling events. asm.org For example, this compound has been shown to inhibit MIP-1β-stimulated γ-S-GTP binding, indicating inhibition of chemokine-dependent stimulation of GDP-GTP exchange at the CCR5/G protein complex. asm.org It also inhibits the downstream event of chemokine-induced intracellular calcium redistribution. asm.org Importantly, this compound does not trigger the release of intracellular calcium, indicating a lack of CCR5 agonist activity. asm.org While this compound blocks chemokine binding and signaling, this is not the primary mechanism by which it exerts its antiviral activity. pmda.go.jp The blockade of chemokine binding by this compound can lead to an increase in circulating levels of CCR5 ligands, as receptor-ligand complexes are typically internalized after binding, and this compound prevents this internalization. ashpublications.org
| Chemokine | IC50 (nM) for Binding Inhibition to CCR5 |
| MIP-1α | 3.3 asm.org |
| MIP-1β | 7.2 asm.org |
| RANTES | 5.2 asm.org |
Inhibition of Endogenous Chemokine Ligand Binding (e.g., MIP-1α, MIP-1β, RANTES)
This compound effectively inhibits the binding of the major endogenous CCR5 ligands, including Macrophage Inflammatory Protein-1 alpha (MIP-1α, also known as CCL3), Macrophage Inflammatory Protein-1 beta (MIP-1β, also known as CCL4), and Regulated upon Activation, Normal T Cell Expressed and Secreted (RANTES, also known as CCL5), to the CCR5 receptor. asm.orgselleckchem.comnih.govdovepress.compmda.go.jpselleck.co.jprndsystems.comuthscsa.edu
Studies using radioligand binding competition assays have quantified the potency of this compound in inhibiting the binding of these chemokines to CCR5-expressing cells or membrane preparations. asm.orgselleckchem.comnih.govpmda.go.jpselleck.co.jp The half-maximal inhibitory concentrations (IC50) demonstrate high affinity binding of this compound to CCR5, effectively competing with these natural ligands. asm.orgselleckchem.comnih.govdovepress.compmda.go.jpselleck.co.jputhscsa.edu
| Chemokine Ligand | IC50 (nM) | 95% Confidence Interval (nM) | Reference |
| MIP-1α (CCL3) | 3.3 | 1.9 to 5.7 | asm.orgselleckchem.comnih.govdovepress.compmda.go.jpselleck.co.jp |
| MIP-1β (CCL4) | 7.2 | 5.5 to 9.5 | asm.orgselleckchem.comnih.govdovepress.compmda.go.jpselleck.co.jp |
| RANTES (CCL5) | 5.2 | 2.1 to 13 | asm.orgselleckchem.comnih.govdovepress.compmda.go.jpselleck.co.jp |
These findings indicate that this compound's binding to CCR5 prevents the natural chemokines from occupying the receptor, thereby blocking their normal function. asm.orgselleckchem.comnih.govpmda.go.jp The inhibition of chemokine binding is reported to be insurmountable under certain conditions, suggesting that circulating chemokine concentrations are unlikely to significantly affect this compound's binding to CCR5. pmda.go.jp
Effects on Downstream CCR5 Signaling Pathways
Beyond blocking ligand binding, this compound also inhibits the downstream signaling events typically triggered by chemokine activation of CCR5. asm.orgselleckchem.comnih.govdovepress.compmda.go.jpchemicalbook.com CCR5 is a G protein-coupled receptor (GPCR), and its activation by chemokines initiates a cascade of intracellular signaling events. nih.govasm.orgselleckchem.comscirp.org this compound, acting as a functional antagonist or inverse agonist, prevents these signaling cascades. asm.orgselleckchem.comnih.gov
Modulation of G-Protein Coupling and GDP-GTP Exchange
Activation of CCR5 by its chemokine ligands leads to the coupling of the receptor with heterotrimeric G proteins and the subsequent exchange of GDP for GTP on the Gα subunit. nih.govasm.orgselleckchem.comxcessbio.commedkoo.com This GDP-GTP exchange is a crucial step in initiating downstream signaling pathways. asm.orgselleckchem.comxcessbio.commedkoo.com
Research has shown that this compound inhibits MIP-1β-stimulated γ-S-GTP binding to cell membranes expressing CCR5, such as HEK-293 cells. asm.orgselleckchem.comxcessbio.commedkoo.com This demonstrates that this compound effectively blocks the chemokine-dependent stimulation of GDP-GTP exchange at the CCR5/G protein complex. asm.orgselleckchem.comxcessbio.commedkoo.com By preventing this initial step of G-protein activation, this compound abrogates the subsequent signaling events. asm.orgselleckchem.comxcessbio.commedkoo.com
Inhibition of Chemokine-Induced Intracellular Calcium Redistribution
One of the key downstream signaling events mediated by CCR5 activation is the rapid redistribution of intracellular calcium, often referred to as calcium flux. asm.orgselleckchem.compmda.go.jpmedkoo.comnih.govresearchgate.netresearchgate.netmedchemexpress.com This increase in intracellular calcium is a common indicator of GPCR activation and plays a role in various cellular responses, including chemotaxis. asm.orgselleckchem.compmda.go.jpmedkoo.comnih.govresearchgate.netresearchgate.netmedchemexpress.com
This compound has been shown to inhibit chemokine-induced intracellular calcium redistribution in a dose-dependent manner. asm.orgselleckchem.compmda.go.jpmedkoo.comnih.govresearchgate.netresearchgate.netmedchemexpress.com Studies using calcium-sensitive dyes and fluorescence assays have quantified this inhibitory effect against various chemokines, including MIP-1α, MIP-1β, and RANTES. asm.orgselleckchem.compmda.go.jpmedkoo.comnih.govresearchgate.netresearchgate.netmedchemexpress.com
| Chemokine Ligand | IC50 (nM) Range (Calcium Redistribution Inhibition) | Reference |
| MIP-1β | 7 to 30 | asm.orgselleckchem.compmda.go.jpmedkoo.commedchemexpress.com |
| MIP-1α | 7 to 30 | asm.orgselleckchem.compmda.go.jpmedkoo.commedchemexpress.com |
| RANTES | 7 to 30 | asm.orgselleckchem.compmda.go.jpmedkoo.commedchemexpress.com |
Importantly, at concentrations up to 10 µM, this compound itself does not trigger the release of intracellular calcium, confirming its lack of agonist activity on CCR5. asm.orgselleckchem.com
Effects on CCR5 Receptor Cell Surface Expression and Internalization
The interaction of chemokine ligands with CCR5 typically leads to the internalization of the receptor from the cell surface, a process that helps regulate the cell's responsiveness to chemokines. asm.orgpmda.go.jpresearchgate.netoup.comoup.comfrontiersin.orgashpublications.org this compound's effect on CCR5 cell surface expression and internalization has been investigated.
Studies have shown that this compound does not induce CCR5 internalization. asm.orgselleckchem.comnih.govnih.gov Flow cytometry experiments have demonstrated that this compound treatment does not lead to a reduction in CCR5 cell surface levels. asm.orgselleckchem.comresearchgate.netresearchgate.net This is consistent with its role as a functional antagonist rather than an agonist. asm.orgselleckchem.comnih.govresearchgate.net
Viral Tropism and Maraviroc Efficacy in Research Settings
Determinants of HIV-1 Co-receptor Usage (V3 Loop)
The third hypervariable loop (V3 loop) of the HIV-1 gp120 protein is recognized as the principal determinant of viral tropism and co-receptor specificity. asm.orgnih.govtandfonline.comresearchgate.netoup.complos.org This region, typically 35 to 37 amino acids in length, plays a crucial role in binding to either CCR5 or CXCR4 after gp120 has bound to CD4. asm.orgresearchgate.netplos.org
Furthermore, structural disorder within the V3 loop may contribute to the switch in cell tropism. Research indicates that the V3 loop of CXCR4-tropic viruses may have a significantly higher intrinsic disorder tendency compared to CCR5-tropic viruses. plos.org
Specificity of Maraviroc for CCR5-Tropic HIV-1 Variants
This compound is a CCR5 antagonist, a class of antiretroviral drugs that specifically block the CCR5 co-receptor, thereby preventing HIV-1 entry into host cells that utilize CCR5. researchgate.net Due to its mechanism of action, this compound is only effective against HIV-1 variants that exclusively use CCR5 for entry (CCR5-tropic or R5 viruses). researchgate.net
Research has consistently demonstrated the specificity of this compound for CCR5-tropic HIV-1. Studies evaluating the efficacy of this compound have focused on patient populations harboring R5 viruses. The presence of CXCR4-using variants (CXCR4-tropic or X4, and dual/mixed-tropic or D/M viruses) confers resistance to this compound because these viruses can utilize CXCR4 for entry even when CCR5 is blocked. Therefore, determining viral tropism before initiating this compound therapy is crucial for predicting treatment response.
Methodologies for Research-Based Viral Tropism Determination
Accurate determination of HIV-1 tropism is essential in research settings to understand viral pathogenesis, study the evolution of tropism, and evaluate the potential efficacy of co-receptor antagonists like this compound. Several methodologies have been developed for research-based viral tropism determination, broadly categorized into phenotypic and genotypic assays. scielo.br
Phenotypic Tropism Assays
Phenotypic assays are cell-based methods that directly assess the ability of a patient's viral population to infect cell lines expressing either CCR5 or CXCR4, or both. scielo.brjmicrobiol.or.kr These assays involve co-culturing patient samples (e.g., peripheral blood mononuclear cells or plasma) with indicator cell lines engineered to express CD4 and either CCR5 or CXCR4. Viral replication or entry into these cell lines is then measured, typically by detecting viral proteins (like p24 antigen) or reporter gene expression.
A widely used phenotypic assay is the Trofile assay. plos.org This assay utilizes recombinant viruses pseudotyped with the patient's env gene, allowing for the evaluation of viral entry into CCR5- and CXCR4-expressing cell lines. The outcome indicates whether the dominant viral population is R5, X4, or D/M. Phenotypic assays are considered the gold standard for tropism determination as they reflect the functional ability of the virus to use a specific co-receptor. However, they are generally more complex, time-consuming, and expensive than genotypic methods. plos.orgscielo.br
Genotypic Tropism Assays (V3 Loop Sequencing)
Genotypic assays infer viral tropism by analyzing the genetic sequence of the V3 loop of the HIV-1 env gene. plos.orgscielo.br Since the V3 loop is the primary determinant of tropism, specific amino acid sequences or patterns within this region can be used to predict co-receptor usage. scielo.br
Traditional genotypic assays involve sequencing the V3 loop region from viral RNA extracted from patient plasma or proviral DNA from peripheral blood mononuclear cells. The resulting sequences are then analyzed using bioinformatic algorithms and rules that correlate specific sequence features (such as the presence of positively charged amino acids at key positions) with predicted tropism. plos.org The "11/25 rule," which associates a positive charge at amino acid positions 11 or 25 with CXCR4 usage, is a historical example of such a rule. plos.orgscielo.br More sophisticated algorithms have been developed that consider multiple positions and physicochemical properties of the V3 loop to improve prediction accuracy. plos.orgjmicrobiol.or.kr
Genotypic assays are generally faster, less expensive, and more widely available than phenotypic assays. However, their predictive accuracy can be influenced by the complexity of the viral population and the specific algorithm used for interpretation.
Advanced Genetic Sequencing Technologies (e.g., Ultra-Deep Pyrosequencing)
Advanced genetic sequencing technologies, such as ultra-deep pyrosequencing (now often referred to as next-generation sequencing or deep sequencing), have revolutionized the ability to detect minority viral variants within a patient's viral population. These technologies can sequence millions of individual DNA or RNA molecules, providing a highly sensitive snapshot of the viral quasispecies.
Applied to tropism testing, deep sequencing of the V3 loop allows for the identification of low-abundance CXCR4-using variants that might be missed by traditional sequencing methods. The presence of even a small population of X4 variants (e.g., above a certain detection threshold, often 0.5% to 2%) can impact the response to CCR5 antagonists like this compound. Therefore, deep sequencing offers a more comprehensive assessment of viral tropism, particularly in the context of predicting response to this compound, by revealing the presence of minority X4 populations that could lead to treatment failure.
Research on Predictive Capacity of Tropism Assays for this compound Efficacy
Extensive research has been conducted to evaluate the predictive capacity of both phenotypic and genotypic tropism assays for the efficacy of this compound. These studies are crucial for guiding clinical decision-making and understanding the implications of viral tropism for treatment outcomes.
Studies have consistently shown that a negative test for CXCR4-using virus by phenotypic or sensitive genotypic assays is a strong predictor of virological response to this compound therapy. Conversely, the presence of detectable CXCR4-using virus, even at low levels, is associated with a significantly reduced likelihood of achieving virological suppression on a this compound-containing regimen.
Research comparing different tropism assay methodologies has highlighted the increased sensitivity of deep sequencing for detecting minority X4 variants compared to traditional sequencing. This increased sensitivity has been shown to improve the prediction of this compound treatment outcomes. For instance, studies have demonstrated that detecting minority X4 populations by deep sequencing is associated with a higher risk of this compound failure compared to results from less sensitive genotypic or phenotypic assays.
Data from clinical trials and observational studies have provided valuable insights into the predictive value of tropism testing. For example, analyses of data from the MOTIVATE studies (Phase 3 trials of this compound) demonstrated that baseline tropism determined by the enhanced sensitivity Trofile assay was a strong predictor of virological response. Patients with R5-tropic virus experienced significantly higher rates of virological success compared to those with D/M tropism. Research using deep sequencing in these and other cohorts has further refined the understanding of the impact of low-level X4 variants on this compound outcomes.
While phenotypic assays like Trofile have been considered the gold standard, the increased sensitivity, speed, and decreasing cost of deep sequencing have made genotypic assays, particularly those utilizing advanced sequencing technologies, increasingly important in research and clinical practice for predicting this compound efficacy. Research continues to refine bioinformatic algorithms for interpreting V3 loop sequences from deep sequencing data to optimize the prediction of co-receptor usage and, consequently, the likelihood of response to CCR5 antagonists.
| Assay Type | Methodology | Advantages | Disadvantages | Predictive Capacity for this compound Efficacy (Research Context) |
| Phenotypic Assays | Cell-based infection assays | Reflects functional tropism; Gold standard | Complex, time-consuming, expensive | Strong predictor; Enhanced sensitivity assays improve prediction |
| Genotypic Assays (Traditional Sequencing) | V3 loop sequencing + bioinformatic analysis | Faster, less expensive, more widely available | Lower sensitivity for minority variants | Predictive, but may miss minority X4 leading to treatment failure |
| Genotypic Assays (Deep Sequencing) | Ultra-deep V3 loop sequencing + analysis | High sensitivity for minority variants; Faster than phenotypic | Requires specialized equipment and expertise; Bioinformatic challenges | Improved prediction by detecting minority X4 variants |
Table 1: Comparison of Research-Based Viral Tropism Determination Methodologies
| Study/Finding | Key Research Finding Related to this compound Efficacy | Relevant Tropism Assay Methodologies |
| Analysis of MOTIVATE studies | Baseline R5 tropism by enhanced sensitivity phenotypic assay strongly predicted virological response to this compound. | Phenotypic (Enhanced Sensitivity) |
| Research using Deep Sequencing | Detection of minority X4 variants (e.g., >0.5-2%) associated with reduced response or failure on this compound. | Genotypic (Deep Sequencing) |
| Comparison of Genotypic vs. Phenotypic Assays | Deep sequencing shows improved sensitivity for detecting minority X4, leading to better prediction of this compound outcome. | Phenotypic, Genotypic (Traditional & Deep Sequencing) |
Table 2: Examples of Research Findings on Predictive Capacity of Tropism Assays
Mechanisms of Hiv-1 Resistance to Maraviroc
Emergence of CXCR4-Using Viruses
One significant mechanism of Maraviroc resistance is the emergence of viruses that utilize the CXCR4 coreceptor for entry, either exclusively (X4-tropic) or in addition to CCR5 (dual/mixed-tropic). asm.orgoup.comscientificarchives.com This shift in coreceptor usage renders the virus insensitive to this compound, which specifically targets CCR5. asm.orgoup.com
Selection of Pre-existing Minority CXCR4-Tropic Populations
The emergence of CXCR4-using viruses during this compound therapy is often attributed to the selection and outgrowth of pre-existing minor CXCR4-tropic variants that were present in the patient's viral population before treatment initiation. asm.orgscientificarchives.comasm.orgdovepress.complos.orgnih.govdaig-net.denih.gov These minority variants, typically present at low frequencies, gain a selective advantage when the CCR5-tropic population is suppressed by this compound. asm.orgasm.orgdovepress.comdaig-net.de Studies have shown that in patients experiencing virologic failure on this compound, the emergent CXCR4-using virus often originates from a pretreatment reservoir rather than a de novo coreceptor switch. asm.orgplos.orgnih.govnatap.org
Phylogenetic Analysis of Tropism Evolution Under Selective Pressure
Phylogenetic analysis of viral envelope (Env) sequences from samples taken before and during this compound treatment has provided insights into the evolution of tropism under drug pressure. asm.orgplos.orgasm.orgplos.org These analyses generally support the hypothesis that emergent CXCR4-using variants are closely related to pre-existing CXCR4-using strains, indicating selection rather than mutation-driven switching from a CCR5-tropic population. asm.orgplos.orgnatap.orgasm.org While de novo coreceptor switching can occur, it is considered less common in patients treated with this compound and typically requires the accumulation of multiple mutations. oup.comscientificarchives.comdovepress.complos.org
Mutations Allowing Use of this compound-Bound CCR5
Another mechanism of this compound resistance involves the development of mutations in the viral Env protein, primarily in gp120, that enable the virus to utilize CCR5 even when it is bound by this compound. asm.orgoup.comscientificarchives.com This is known as non-competitive resistance and is characterized by a reduction in the maximal percentage of viral inhibition by this compound. dovepress.comoup.comresearchgate.netresearchgate.netasm.orgasm.org
Role of gp120 Mutations, Particularly within the V3 Loop
Mutations in the gp120 protein, particularly within the V3 loop, play a crucial role in allowing HIV-1 to use this compound-bound CCR5. asm.orgdovepress.comresearchgate.netresearchgate.netasm.orgasm.orgresearchgate.netplos.orgfrontiersin.orgasm.orgnih.govdntb.gov.uaescholarship.org The V3 loop is a key determinant of coreceptor tropism and is involved in the interaction with the extracellular loops of CCR5. plos.orgasm.orgnih.govfrontiersin.org Mutations in this region can alter the conformation and dynamics of the V3 loop, enabling it to interact with the drug-bound state of CCR5. plos.orgnih.gov While V3 loop mutations are commonly associated with this type of resistance, mutations outside the V3 loop, such as in the C4 region, have also been implicated. researchgate.netasm.orgnih.gov
Altered Affinity of gp120 for Inhibitor-Bound CCR5
Resistance mutations can lead to an altered affinity of gp120 for CCR5 in the presence of this compound. dovepress.comresearchgate.netasm.orgresearchgate.netasm.org While this compound binding induces conformational changes in CCR5, particularly in the extracellular loops, resistant viruses develop the ability to recognize and efficiently engage with this drug-bound conformation. dovepress.comresearchgate.netasm.orgasm.orgnih.gov This can involve increased binding to specific CCR5 domains that are less affected by this compound binding. dovepress.comresearchgate.netnih.gov The level of resistance is related to the virus's relative affinity for inhibitor-bound versus drug-free CCR5. asm.org
Involvement of CCR5 N-terminal and Extracellular Loop Domains in Resistance
The interaction between gp120 and CCR5 involves multiple domains of the coreceptor, including the N-terminal domain and the extracellular loops (ECLs). asm.orgasm.orgresearchgate.netasm.orgnih.govfrontiersin.org this compound primarily affects the conformation of the ECLs. dovepress.comresearchgate.netasm.orgnih.govnih.gov However, resistant viruses can become critically dependent on the CCR5 N-terminus and specific residues within the drug-modified ECLs for entry. asm.orgasm.orgresearchgate.netnih.gov This suggests that resistance can involve complex interactions with both the N-terminal and ECL domains of this compound-bound CCR5, and the specific interactions may differ depending on the viral strain and resistance mutations. asm.orgasm.orgresearchgate.netescholarship.orgnih.gov
Compound Table
| Compound Name | PubChem CID |
| This compound | 3002977 nih.govmims.comnih.govgpcrmd.orgguidetopharmacology.org |
Data Tables
Based on the provided text, specific quantitative data suitable for interactive tables is limited. However, the following conceptual table summarizes the two primary mechanisms of resistance:
| Mechanism of Resistance | Description | Viral Population Involved | Key Viral/Host Factors Involved |
| Emergence of CXCR4-Using Viruses | Selection and outgrowth of pre-existing CXCR4-tropic or dual/mixed-tropic HIV-1 variants. | Pre-existing minority CXCR4-using variants. | HIV-1 Env (gp120) tropism determinants, CCR5 and CXCR4 coreceptors. |
| Mutations Allowing Use of this compound-Bound CCR5 | Development of mutations in viral Env (primarily gp120) allowing entry via this compound-occupied CCR5. | CCR5-tropic HIV-1 acquiring resistance mutations. | HIV-1 gp120 (especially V3 loop), CCR5 N-terminus and ECL domains. |
This table provides a structured overview of the two main resistance pathways discussed in the article. More detailed quantitative data, such as specific mutation frequencies or changes in binding affinities, were described qualitatively in the source snippets rather than in discrete data sets suitable for automated interactive table generation without further processing and interpretation.
In Vitro Selection of this compound-Resistant Variants
In vitro studies have been instrumental in understanding how HIV-1 develops resistance to this compound. These experiments often involve exposing viral isolates to increasing concentrations of the drug over time.
Serial passage experiments, where HIV-1 is cultured in the presence of escalating concentrations of this compound, have demonstrated the potential for resistance to emerge tga.gov.auasm.org. While some laboratory-adapted and clinical isolates failed to select for this compound resistance under these conditions, high-level resistance was successfully selected from some primary isolates passaged in peripheral blood lymphocytes (PBLs) asm.orgnih.gov.
For instance, high-level resistance was selected from three out of six primary isolates in one study asm.orgnih.gov. Interestingly, one strain, SF162, acquired resistance in both treated and control cultures, with resistant variants becoming CXCR4-using asm.orgnih.gov. In contrast, this compound-resistant virus derived from isolates like CC1/85 and RU570 remained CCR5-tropic, retaining susceptibility to other CCR5 antagonists like SCH-C and resistance to CXCR4 antagonists like AMD3100 asm.orgnih.gov. These resistant variants showed an inability to replicate in CCR5 Δ32/Δ32 PBLs, further confirming their CCR5 tropism asm.orgnih.gov.
Phenotypic resistance to this compound in R5 viruses is often characterized by a reduced maximal percentage inhibition (MPI) in dose-response curves, rather than a significant shift in the half-maximal inhibitory concentration (IC50) dovepress.comnatap.orgnatap.orgplos.org. This indicates that resistant viral strains can utilize the CCR5 co-receptor even when this compound is bound dovepress.comnatap.orgasm.orgplos.org.
In phenotypic susceptibility assays, a reduced MPI suggests that the virus has acquired the ability to utilize the this compound-bound receptor for entry dovepress.comasm.orgnih.gov. Most resistant viruses typically exhibit an MPI between 80% and 95% dovepress.com. Studies analyzing viruses from patients who experienced virologic failure while on this compound have validated the use of reduced MPI as a phenotypic marker of resistance in vivo natap.org. For example, in the MOTIVATE trials, plateaus in MPI below 95% were identified in some patients failing this compound therapy with R5 virus natap.orgnatap.orgplos.org. Clonal analyses further confirmed that Env clones from these failure time points displayed reduced MPI plos.org.
Serial Passage Experiments and Resistance Development
Cross-Resistance Patterns with Other CCR5 Antagonists
Resistance to one CCR5 antagonist does not always result in broad cross-resistance to other agents in the same class asm.orgresearchgate.netnih.gov. While resistance to some CCR5 antagonists can lead to broad cross-resistance, this compound-resistant viruses have been observed to have narrow cross-resistance patterns asm.orgresearchgate.net.
Studies analyzing viruses from patients failing this compound-containing regimens have shown that while high-level resistance to this compound can develop, these viruses may remain sensitive to most other CCR5 antagonists, including vicriviroc and aplaviroc asm.orgresearchgate.netnih.gov. Some this compound-resistant variants have shown partial cross-resistance to compounds like TAK779 but remained sensitive to others researchgate.net. This suggests that the specific mechanisms of resistance can influence the degree of cross-resistance to different CCR5 antagonists asm.org.
Genetic and Structural Basis of this compound Resistance
The development of this compound resistance in R5 viruses is primarily associated with genetic changes in the viral envelope glycoprotein gp120, particularly in the V3 loop region oup.comasm.orgnatap.orgtga.gov.aunatap.orgplos.orgresearchgate.net. These mutations enable the virus to interact with and utilize the CCR5 co-receptor even when this compound is bound dovepress.comoup.comasm.org.
Mutations associated with this compound resistance are frequently found in the V3 loop of gp120 natap.orgtga.gov.auasm.orgnatap.orgplos.orgasm.orgresearchgate.net. However, there is a notable lack of a single, predictable set of "signature" mutations that consistently confer resistance across different patients or isolates asm.orgnatap.orgplos.org. The specific amino acid changes can differ between patients, reflecting the heterogeneity of the gp160 sequence natap.org. While V3 loop mutations are key determinants natap.orgasm.orgnih.govplos.orgasm.org, mutations outside the V3 loop, in other regions of gp120 or even in gp41, can also contribute to resistance asm.orgresearchgate.net.
Structural studies and modeling suggest that this compound binds deep within a hydrophobic pocket in the transmembrane region of CCR5, inducing conformational changes in the extracellular loops (ECLs) dovepress.comresearchgate.netrcsb.orgnih.gov. Resistance mutations in gp120, particularly in the V3 loop, are thought to alter the interaction between gp120 and the drug-bound CCR5 dovepress.comoup.comasm.orgnih.govplos.org. These mutations may lead to increased affinity of gp120 for the CCR5-maraviroc complex, allowing entry despite the conformational changes induced by the drug dovepress.com. Some studies indicate that resistant viruses may show altered reliance on different domains of CCR5, such as increased dependence on the N-terminal domain compared to the ECLs, although this compound-resistant viruses with narrow cross-resistance may still depend on drug-modified ECLs asm.orgasm.org. Molecular dynamics simulations of the gp120 V3 loop with resistance mutations have shown altered V3 loop structure and dynamics, which could facilitate interaction with drug-bound CCR5 plos.org.
The complex and variable nature of the genetic changes associated with this compound resistance presents a challenge for the development of simple genotypic resistance algorithms natap.orgplos.org.
Table 1: Selected Mutations Associated with this compound Resistance
| gp120 Region | Mutation Example(s) (HXB2 numbering) | Reference | Notes |
| V3 Loop | T316A, V323I | asm.orgnih.gov | Reversion restored sensitivity; single mutations resulted in partial resistance. |
| V3 Loop | A316T, I323V, S405A | dovepress.com | Observed in some patients failing treatment. |
| V3 Loop | P/T308H, H320T, V322aI | dovepress.com | Associated with varying degrees of resistance. |
| V3 Loop | I304V, F312W, T314A, E317D, I318V | plos.org | Combinations required for resistance in one study. |
| V3 Loop | Various (patient-specific) | natap.orgplos.org | No consistent pattern observed across patients. |
| V4 Loop | Various | asm.orgnih.gov | Modulated the magnitude of resistance in one study. |
| gp41 | Mutations in fusion peptide | researchgate.net | In vitro evidence suggesting potential role. |
Table 2: Phenotypic Characteristics of this compound Resistance
| Phenotypic Marker | Description | Observation in Resistant Viruses | Reference |
| Maximal Percentage Inhibition (MPI) | The maximum level of viral inhibition achieved by the drug. | Reduced MPI (typically 80-95%), indicating incomplete inhibition. | dovepress.comnatap.orgnatap.orgplos.org |
| IC50 Shift | Change in the drug concentration required for 50% inhibition. | Often minimal or inconsistent; reduced MPI is a more reliable marker. | natap.orgnatap.orgplos.org |
| Utilization of Drug-Bound CCR5 | Ability of the virus to enter cells via CCR5 even when this compound is bound. | This is the underlying mechanism for reduced MPI. | dovepress.comoup.comnatap.orgasm.orgplos.org |
Table 3: Cross-Resistance of this compound-Resistant Viruses
| Other CCR5 Antagonist | Susceptibility of this compound-Resistant Virus | Reference | Notes |
| Vicriviroc | Sensitive | asm.orgresearchgate.netnih.gov | Observed in some this compound-resistant isolates. |
| Aplaviroc | Sensitive | asm.orgresearchgate.netnih.gov | Observed in some this compound-resistant isolates. |
| TAK779 | Partially cross-resistant | researchgate.net | Variable findings depending on the resistant strain. |
| SCH-C | Sensitive | asm.orgnih.govaidsmap.com | Observed in some in vitro selected variants. |
| SCH-D (Vicriviroc) | Sensitive | aidsmap.com | Observed in some in vitro selected variants. |
In Vitro Antiviral Activity and Synergism Studies
Potency Against Diverse CCR5-Tropic HIV-1 Isolates (various clades and geographic origins)
Maraviroc has demonstrated potent in vitro antiviral activity against a broad spectrum of CCR5-tropic HIV-1 viruses. Studies involving 43 primary isolates from various clades and diverse geographic origins have shown a geometric mean 90% inhibitory concentration (IC90) of approximately 2.0 nM. The in vitro IC50 (50% inhibitory concentration) for this compound against the replication of HIV-1 group M isolates (subtypes A to J and circulating recombinant form AE) and group O isolates has been reported to range from 0.1 to 4.5 nM (0.05 to 2.3 nanogram/mL) in cell culture. This indicates consistent potency across different major subtypes and geographic sources of CCR5-tropic HIV-1. No single exclusively CCR5-tropic subtype of HIV-1 appeared to have significantly lower susceptibility to this compound than others, although one hybrid subtype (CRF_01(AE)) showed markedly greater susceptibility.
Activity Against HIV-1 Strains Resistant to Other Antiretroviral Classes
A significant finding from in vitro research is this compound's activity against HIV-1 strains that have developed resistance to other classes of antiretroviral agents. HIV-1 clinical isolates resistant to nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), protease inhibitors (PIs), and enfuvirtide have all shown susceptibility to this compound in cell culture. This lack of cross-resistance with existing agents that target intracellular steps is a key advantage of entry inhibitors like this compound. Studies using clinically derived HIV-1 envelope pseudoviruses, including those from viruses resistant to existing drug classes, also confirmed this compound's activity.
Synergistic Antiviral Effects with Other Antiretroviral Agents in Cell Culture
The combination of this compound with other antiretroviral agents in cell culture has been investigated to understand potential synergistic or additive antiviral effects. Generally, additive or slightly synergistic interactions have been observed when this compound is tested in combination with licensed antiretroviral agents from different classes. This supports the use of this compound in multi-drug regimens for HIV-1 infection.
Combinations with Protease Inhibitors
When combined with protease inhibitors (PIs) in vitro, this compound has shown additive to synergistic antiviral effects. Specific PIs studied in combination with this compound include amprenavir, atazanavir, indinavir, lopinavir, nelfinavir, ritonavir, and saquinavir. Moderate synergy was observed in some experiments with atazanavir, indinavir, and nelfinavir, with additive effects seen in repeat experiments or with other PIs.
Combinations with Nucleoside Reverse Transcriptase Inhibitors (NRTIs)
Combinations of this compound with nucleoside reverse transcriptase inhibitors (NRTIs) have generally resulted in additive antiviral effects in cell culture. NRTIs tested in combination with this compound include abacavir, didanosine, emtricitabine, lamivudine, stavudine, tenofovir, zalcitabine, and zidovudine.
Combinations with Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs)
In vitro studies combining this compound with non-nucleoside reverse transcriptase inhibitors (NNRTIs) have indicated generally additive antiviral interactions. NNRTIs evaluated include delavirdine, efavirenz, and nevirapine. Minor synergy was observed with efavirenz in some studies.
Combinations with HIV Fusion Inhibitors (e.g., Enfuvirtide)
This compound has shown additive to synergistic effects when combined with the HIV fusion inhibitor enfuvirtide in cell culture. Moderate synergy was observed in some experiments with enfuvirtide, with additive effects seen in repeat experiments. The synergy between these two classes of entry inhibitors, targeting different steps of the entry process, can be influenced by factors such as the strength of binding between enfuvirtide and viral gp41 and the density of CCR5 receptors on host cells.
Here is a summary of the in vitro synergy results with different antiretroviral classes:
| Antiretroviral Class | Specific Agents Tested | Observed Interaction with this compound (In Vitro) |
| Protease Inhibitors (PIs) | Amprenavir, Atazanavir, Indinavir, Lopinavir, Nelfinavir, Ritonavir, Saquinavir | Additive to Synergistic |
| Nucleoside Reverse Transcriptase Inhibitors (NRTIs) | Abacavir, Didanosine, Emtricitabine, Lamivudine, Stavudine, Tenofovir, Zalcitabine, Zidovudine | Generally Additive |
| Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs) | Delavirdine, Efavirenz, Nevirapine | Generally Additive (Minor Synergy observed with Efavirenz) |
| HIV Fusion Inhibitors | Enfuvirtide | Additive to Synergistic |
Structural Biology and Computational Modeling of Maraviroc-ccr5 Interactions
CCR5 Receptor Structural Characterization and Binding Pocket Analysis
The CCR5 receptor, like other GPCRs, is characterized by a structure comprising seven transmembrane (7TM) α-helices connected by intracellular and extracellular loops. ulb.ac.beacs.orgwikipedia.org The first high-resolution crystal structure of human CCR5 bound to Maraviroc (PDB ID: 4MBS) was reported in 2013, providing crucial insights into the receptor's architecture and ligand binding. scirp.org This structure revealed a ligand binding site located within the 7TM bundle. acs.orgnih.gov
The ligand binding pocket of CCR5 is described as a cavity within the transmembrane domain. wikipedia.orgnih.gov This pocket is notably hydrophobic and lined with multiple aromatic residues. wikipedia.org Compared to other class A GPCRs, the 7TM ligand binding pockets of chemokine receptors like CCR5 are less buried and more solvent accessible. acs.org The extracellular loop 2 (ECL2) covers the pocket to a lesser extent than in most other GPCRs, which is thought to be related to chemokine receptors binding larger chemokine ligands. acs.org
Site-directed mutagenesis and biochemical studies have indicated that small molecule CCR5 inhibitors, including this compound, bind to a hydrophobic cavity within the transmembrane domain. nih.gov This location is distinct from the proposed major binding sites for endogenous chemokines and the viral glycoprotein gp120, which are located at the receptor surface, particularly involving the N-terminus and ECL2. nih.govnih.gov
Role of Transmembrane Helices and Extracellular Loops in Ligand Binding
The transmembrane helices and extracellular loops of CCR5 play critical roles in ligand binding and receptor function. The 7TM bundle forms the core of the receptor and contains the binding site for small molecule antagonists like this compound. acs.orgwikipedia.orgnih.gov Specific residues within these transmembrane helices contribute significantly to ligand binding. researchgate.netnih.gov For instance, studies on the binding of the antagonist TAK779 to CCR5 suggest interactions with residues in helices 1, 2, 3, and 7, with additional interactions with helices 5 and 6. nih.gov
The extracellular domains, particularly the N-terminus and extracellular loops (ECLs), are crucial for the interaction with chemokine ligands and the HIV-1 envelope protein gp120. ulb.ac.benih.govresearchgate.netoup.com The N-terminal domain has been shown to be mandatory for chemokine binding, with charged and aromatic residues playing a key role. ulb.ac.be The second extracellular loop (ECL2) is also important for ligand specificity and interaction with the V3 loop of gp120. ulb.ac.benih.govoup.complos.org While chemokines and gp120 interact with these extracellular regions, small molecule antagonists like this compound primarily interact with the transmembrane domain binding pocket. acs.orgnih.govnih.gov
This compound Binding Site and Specificity Analysis
The crystal structure of the CCR5-Maraviroc complex revealed that this compound is buried in a cavity within the 7TM domain. nih.gov This binding site is distinct from the primary recognition sites for chemokines and gp120. nih.gov this compound is predicted to insert deeply into the CCR5 transmembrane cavity, potentially occupying different binding sites. nih.gov This location suggests that this compound inhibits chemokine function by blocking receptor activation through interactions with a site that potentially overlaps with the chemokine activation site (Site 2), located within the 7TM domain and interacting with the flexible N-terminus of the chemokine. nih.govresearchgate.net
This compound's binding to CCR5 is characterized by its specificity. It functions as an antagonist of CCR5, which is the main co-receptor for R5-tropic HIV-1 strains. nih.govnih.gov The binding site for this compound has been shown to be distinct from that of other CCR5 antagonists like TAK-779, despite some overlap in key residues involved in binding. natap.org Differences in the specific interactions within the binding site can lead to variations in the conformational changes induced and the resulting resistance profiles against different antagonists. natap.org
Conformational Dynamics and Allosteric Shifts Induced by this compound Binding
This compound is characterized as an inverse agonist of CCR5, suggesting that its binding stabilizes the receptor in an inactive conformation. nih.gov This is supported by the conformation of conserved GPCR residues, such as Trp248 and Tyr244, which are involved in transmitting ligand-induced conformational changes. nih.gov In the CCR5-Maraviroc structure, these residues adopt conformations similar to those observed in other inactive GPCR structures. nih.gov
This compound binding to CCR5 provokes an allosteric conformational change that inhibits viral engagement with the co-receptor and prevents entry. nih.gov This allosteric mechanism involves this compound binding to a site distinct from the orthosteric site where chemokines and gp120 bind, yet influencing the conformation of these binding sites. nih.govnih.govmdpi.comasm.org The binding of small molecules like this compound is associated with compound-specific conformational changes in the binding site. natap.org These changes are speculated to translate to compound-specific alterations in the extracellular loops, which in turn modulate the binding site for HIV gp120, preventing viral docking. natap.org Studies have shown that this compound induces conformational changes in the CCR5 receptor to block HIV interaction while still allowing native chemokine internalization of the receptor. mdpi.com
Specific residues are involved in the binding and the induced conformational changes. Key residues for the initial binding of both this compound and another CCR5 inhibitor, RO1752, include W86 and I198. natap.org The residue F85 is involved in the conformational change to the final stable state for both compounds. natap.org However, the conformational change induced by this compound is differentiated by the contribution of Y108 and Y251 to its binding. natap.org Mutation of residues Y108 on helix III and Y251 on helix VI to alanine eliminated the time-dependent binding for this compound, but not for RO1752, indicating distinct dynamics in their binding and induced conformational changes. natap.org
Development of Allosteric Models for CCR5 Antagonism
The observation that this compound binds to a site distinct from the chemokine/gp120 binding site and induces conformational changes has led to the development of allosteric models for CCR5 antagonism. nih.govnih.govasm.org These models propose that this compound binding to a transmembrane cavity allosterically prevents the binding of its natural ligands and gp120 by stabilizing an inactive receptor conformation. nih.govnih.gov
Studies combining site-directed mutagenesis and molecular modeling have supported the view that small molecule CCR5 inhibitors bind to a hydrophobic cavity within the transmembrane domain, acting allosterically. nih.gov An allosteric model suggests that this compound enhances the dissociation of preformed ligand-CCR5 complexes, implying that the transmembrane cavity remains accessible even when other ligands are bound. nih.gov
Allosteric antagonism provides a mechanism where the modulator binds to a site topologically distinct from the orthosteric site but still affects receptor function. nih.govfrontiersin.org In the case of CCR5, this compound's allosteric binding stabilizes a receptor conformation that is not recognized by HIV-1, thereby preventing entry. natap.org This is in contrast to competitive inhibition, where the inhibitor directly competes with the natural ligand for the same binding site.
Application of Molecular Docking and Molecular Dynamics Simulations
Computational techniques such as molecular docking and molecular dynamics (MD) simulations have been extensively applied to study the interaction of this compound with CCR5 and to understand the structural basis of its activity. scirp.orgnih.govmdpi.comzenodo.orgtandfonline.com
Molecular docking is used to predict the binding pose and affinity of this compound within the CCR5 binding pocket. scirp.orgnih.govzenodo.org Studies using docking have predicted that this compound inserts deeply into the transmembrane cavity and can occupy multiple binding sites. nih.gov Docking calculations have also been used in virtual screening approaches to identify potential novel CCR5 inhibitors and compare their predicted binding affinities to that of this compound. nih.govmdpi.comzenodo.org For example, some compounds have shown stronger predicted binding affinities than this compound in docking studies, forming favorable interactions with key CCR5 residues like TYR108, GLU283, and PHE109. zenodo.org
Molecular dynamics simulations provide insights into the dynamic behavior of the CCR5-Maraviroc complex, allowing researchers to study the stability of the binding interaction and the conformational changes induced by this compound binding over time. scirp.orgnih.govmdpi.comtandfonline.com MD simulations can predict atomic motions and validate docking results. scirp.org They are used to assess the binding stability of ligands and gain insights into their binding modes. scirp.org MD simulations have been employed to refine CCR5 models, particularly focusing on the flexible extracellular loops and N-terminal segment, and to produce flexible receptor templates for docking studies. plos.org Furthermore, MD simulations, combined with techniques like principal component analysis (PCA) and free energy landscape (FEL) analysis, are used to investigate the conformational dynamics and allosteric mechanisms of CCR5 antagonists. tandfonline.com
Quantitative Structure-Activity Relationship (QSAR) Studies for CCR5 Antagonists
Quantitative Structure-Activity Relationship (QSAR) studies aim to establish a relationship between the chemical structure of a series of compounds and their biological activity, allowing for the prediction of the activity of new compounds and the design of novel inhibitors with improved properties. tandfonline.comrjptonline.orgeurekaselect.comacs.org QSAR studies have been performed on various series of compounds as CCR5 antagonists, including piperidine and piperazine derivatives. tandfonline.comrjptonline.orgeurekaselect.com
These studies utilize various molecular descriptors (structural, spatial, electronic, physicochemical, and topological parameters) and statistical methods (such as multiple linear regression, partial least squares, and artificial neural networks) to build predictive models. tandfonline.comrjptonline.org QSAR models for CCR5 antagonists have revealed that factors such as steric factors and lipophilicities of substituents play a significant role in determining the antagonistic activity. eurekaselect.com The size and shape of substituents are emphasized as important for effective antagonist-CCR5 binding chemistry. eurekaselect.com
Immunological and Cellular Effects of Ccr5 Antagonism by Maraviroc
Modulation of T-Cell Homeostasis and Function in Vitro
In vitro studies have investigated the direct effects of maraviroc on human T lymphocytes, providing insights into how CCR5 blockade influences T-cell behavior. oup.comoup.comresearchgate.net
Effects on T-Cell Activation and Proliferation
In vitro exposure to this compound has been shown to modulate markers of T-cell activation. Studies using peripheral blood mononuclear cells (PBMCs) from both HIV-negative and treated HIV-positive individuals demonstrated that this compound can influence the expression of activation markers such as CD69, CD38, HLA-DR, and CD25. oup.comoup.comresearchgate.netnih.gov While slight differences were observed at high concentrations, the expression of CD25, CD38, and HLA-DR tended to decrease in both CD4+ and CD8+ T lymphocytes, whereas CD69 expression tended to increase. oup.comoup.comresearchgate.netnih.gov
Regarding T-cell proliferation, in vitro studies indicated that this compound tends to inhibit polyclonal-induced proliferation, particularly at higher concentrations (e.g., 100 μM). oup.comoup.comresearchgate.netnih.gov
Influence on CCR5 Expression and Density on T-Cell Surface
This compound has been observed to influence CCR5 expression on the surface of T cells. In vitro studies showed that this compound increases CCR5 surface expression on activated T cells, even at low concentrations (0.1 μM). oup.comoup.comresearchgate.net This effect was observed on both CD4+ and CD8+ T cells after stimulation. oup.comoup.com In non-stimulated PBMCs, CCR5 density also tended to rise in both T-cell subsets when cultured with this compound. oup.comoup.com This increase in CCR5 expression may be a consequence of the disruption of the interaction between CCR5 and its ligands, which typically leads to receptor internalization. plos.org
Inhibition of Chemotaxis of Immune Cells
A significant effect of this compound observed in vitro is the inhibition of immune cell migration. This compound clearly inhibits T-cell migration induced by chemokines in a dose-dependent manner. oup.comoup.comresearchgate.netnih.gov This has been demonstrated for T lymphocytes migrating towards a mixture of chemokines like CCL3, CCL4, and CCL5. oup.comoup.com
Beyond T cells, this compound has also shown the ability to inhibit the chemotactic activity of innate immune cells, including monocytes, macrophages, and monocyte-derived dendritic cells, in response to various chemoattractants such as MIP-1β, MCP-1, and RANTES. nih.govresearchgate.net This inhibition of migration suggests a potential role for this compound in down-regulating inflammation by blocking the trafficking of these cells. nih.gov
Influence on Host Immune Response Beyond Antiviral Activity
The CCR5 antagonism by this compound extends its influence to the host immune response beyond merely blocking viral entry. oup.comashpublications.orgfrontiersin.orgplos.orgnih.govfda.gov
Immunomodulatory Potential of CCR5 Antagonism
CCR5 blockade by this compound has been hypothesized to exert immunomodulatory effects. oup.comfrontiersin.orgnih.govfda.gov These potential effects include the down-regulation of immune activation and changes in cytokine expression. oup.comnih.gov While some studies have indicated that this compound may reduce markers of immune activation, others have found it to increase T-cell activation in certain compartments. ashpublications.orgplos.orgplos.org This suggests that the net immunological benefit or disadvantage on host cellular immune responses can be complex and potentially context-dependent. plos.org
Studies have explored the impact of this compound on T-cell subsets, observing effects such as a decrease in ex-vivo apoptotic CD4+ and CD8+ T cells and a reduction in the loss of CD8+ memory T cells. plos.org this compound has also been shown to potentially reduce the frequency of regulatory T cells (Tregs). dovepress.comasm.org These effects on T-cell populations contribute to the broader immunomodulatory potential attributed to CCR5 antagonism.
Furthermore, the immunomodulatory effects of this compound have been investigated in contexts beyond HIV, such as graft-versus-host disease and potentially in inflammatory processes and certain cancers, where CCR5 plays a role in immune cell recruitment and function. nih.govfrontiersin.orgaacrjournals.orghaematologica.org
Receptor Saturation as a Pharmacodynamic Biomarker
During the development of this compound, CCR5 receptor occupancy (saturation) was investigated as a potential pharmacodynamic biomarker for efficacy. mims.comnih.gov Studies measured the extent to which this compound occupies CCR5 receptors on CD4+ T cells. fda.govnih.gov
Data from clinical trials indicated that dose-dependent saturation of CCR5 receptors was achieved with this compound. nih.govnih.gov Doses of 25 mg once daily and higher resulted in near maximum saturation levels, and receptor saturation remained high for several days after dosing cessation, reflecting slow dissociation from the receptor. nih.gov
However, studies exploring the correlation between the degree of CCR5 receptor saturation and the reduction in viral load found no direct correlation. nih.govresearchgate.net It was suggested that very high levels of receptor saturation might be required for antiviral efficacy, and the variability of the assay at high saturation levels made it difficult to differentiate effectively. nih.gov Consequently, routine monitoring of receptor occupancy was not found to be a helpful biomarker for predicting this compound efficacy in terms of viral load reduction. nih.govresearchgate.net Despite this, receptor saturation measurements were valuable in understanding the binding characteristics and pharmacodynamics of this compound. nih.gov
Compound Names and PubChem CIDs
| Compound Name | PubChem CID |
| This compound | 3002977 |
Interactive Data Tables
Based on the provided text, here are some data points that could be represented in interactive tables:
Table 1: In Vitro Effects of this compound on T-Cell Activation Markers (This table would ideally present changes in expression levels of CD69, CD38, HLA-DR, and CD25 on CD4+ and CD8+ T cells at different this compound concentrations, if specific numerical data were available in the text snippets. The text currently provides directional trends rather than precise values suitable for a table.)
| Activation Marker | T-Cell Subset | This compound Concentration | Observed Effect (Trend) | Source Snippet |
| CD25 | CD4+, CD8+ | High | Decrease | oup.comoup.comresearchgate.netnih.gov |
| CD38 | CD4+, CD8+ | High | Decrease | oup.comoup.comresearchgate.netnih.gov |
| HLA-DR | CD4+, CD8+ | High | Decrease | oup.comoup.comresearchgate.netnih.gov |
| CD69 | CD4+, CD8+ | High | Increase | oup.comoup.comresearchgate.netnih.gov |
Table 2: In Vitro Effect of this compound on Chemokine-Induced T-Cell Migration (This table would ideally show the percentage of migration inhibition at different this compound concentrations. The text mentions dose-dependent inhibition but lacks specific numerical data points for a detailed table.)
| Immune Cell Type | Chemoattractants Used | This compound Concentration | Effect on Migration | Source Snippet |
| T lymphocytes | Mixture of chemokines | Dose-dependent | Inhibition | oup.comoup.comresearchgate.netnih.gov |
| Monocytes | MIP-1β, MCP-1 | Dose-dependent | Significant reduction | nih.gov |
| Monocytes | fMLP | 1 µM and 10 µM | Decreased | nih.gov |
| Macrophages | Tested chemoattractants | 0.1, 1, 10 µM | Significant inhibition | nih.gov |
| Monocyte-derived dendritic cells | Tested chemoattractants | 0.1, 1, 10 µM | Significant inhibition | nih.gov |
Table 3: In Vivo Effects of this compound on CCR5 Expression on T-Cells (Based on Clinical Trial Data) (Based on snippet ashpublications.org, which provides percentage increases in CCR5+ T cells in a clinical trial.)
| T-Cell Subset | Compartment | Change in % CCR5+ Cells (Median/Mean) | Statistical Significance (P-value) | Source Snippet |
| CD8+ | Peripheral blood | +9 percentage points (mean) | < 0.001 | ashpublications.org |
| CD4+ | Peripheral blood | +2.4 percentage points (mean) | 0.012 | ashpublications.org |
| CD4+ | Rectal tissue | Increased nearly twofold | < 0.001 | ashpublications.org |
Table 4: In Vivo Effect of this compound on Plasma CCR5 Ligand Levels (Based on snippet ashpublications.org, which provides the increase in MIP-1β levels in a clinical trial.)
| CCR5 Ligand | Change in Plasma Level (Mean Fold Increase) | Statistical Significance (P-value) | Source Snippet |
| MIP-1β | 2.4-fold | < 0.001 | ashpublications.org |
Interaction with Other Chemokine Receptors and Signaling Pathways
Chemokine receptors are a large family of G protein-coupled receptors (GPCRs) involved in various cellular processes, including immune cell trafficking, inflammation, and cell proliferation. While this compound is designed to specifically target CCR5, understanding its potential interactions with other receptors and downstream signaling cascades is crucial for a comprehensive view of its cellular effects.
Specificity Against Other Chemokine Receptors (e.g., CCR2)
This compound has demonstrated a high degree of selectivity for CCR5. Studies have shown that this compound exhibits no significant activity against a range of other receptors and enzymes, including the hERG ion channel, at concentrations significantly exceeding its half-maximal inhibitory concentrations (IC50s) for CCR5 ligand binding and signaling asm.orgnih.gov.
Of particular note is its selectivity against CCR2, another CC chemokine receptor that shares sequence identity with CCR5 and is known to be susceptible to some other CCR5 antagonists asm.orgnih.govbiorxiv.org. Research indicates that this compound shows no evidence of significant activity against CCR2 asm.orgnih.gov. This specificity is important because CCR2 is involved in the recruitment and migration of monocytes and macrophages, playing a role in various inflammatory and fibrotic conditions tandfonline.commdpi.comwustl.edu. The distinct profiles of CCR2 and CCR5 antagonists, such as the dual CCR2/CCR5 inhibitor cenicriviroc compared to the CCR5-selective this compound, highlight the importance of this specificity in mediating differential cellular and physiological outcomes nih.govtandfonline.comwustl.edu.
The selectivity profile of this compound has been evaluated in human in vitro immune function assays, demonstrating no significant activity against related chemokine receptors at concentrations over 1,000 times the IC50 for this compound's effect on CCR5 asm.org.
Modulation of Downstream Intracellular Signaling Cascades (e.g., JAK/STAT, MAPK/NF-κB)
CCR5, as a GPCR, primarily signals through Gαi proteins upon ligand binding. This can lead to the activation of various downstream intracellular signaling pathways, including those involving calcium mobilization, the PI3K/Akt pathway, and potentially others like JAK/STAT and MAPK/NF-κB asm.orgaacrjournals.org.
While this compound functions as a functional antagonist or inverse agonist of CCR5, blocking chemokine binding and CCR5-mediated signaling, its precise modulation of downstream cascades like JAK/STAT and MAPK/NF-κB in all contexts is an area of ongoing research asm.org. Some studies suggest that CCR5 antagonism can influence these pathways, particularly in the context of inflammatory responses and cell survival aacrjournals.orgnih.gov.
For instance, research in the context of sepsis-associated liver injury suggests that this compound can inhibit the MAPK and NF-κB signaling pathways, thereby reducing inflammation nih.gov. The interconnectedness of signaling pathways like NF-κB and JAK/STAT is recognized, with crosstalk occurring through various mediators, including cytokines like IL-6 mdpi.comresearchgate.netnih.gov. While NF-κB is often associated with pro-inflammatory responses, pathways like PI3K-Akt, which can be influenced by CCR5 signaling, may have anti-inflammatory effects aacrjournals.orgmdpi.com. The modulation of these complex networks by CCR5 antagonism can vary depending on the cell type and the specific biological context aacrjournals.org.
Further detailed research findings on the direct impact of this compound-mediated CCR5 antagonism on JAK/STAT and MAPK/NF-κB signaling in various cell types are crucial for a complete understanding of its cellular mechanisms beyond viral entry inhibition.
Impact on Cell Cycle Regulation in Specific Cell Types
Beyond its role in immune cell function and viral entry, CCR5 signaling has been implicated in regulating cell cycle progression in certain cell types, particularly in the context of cancer. Antagonizing CCR5 with this compound has shown effects on cell proliferation and cell cycle distribution in various cancer cell lines.
Studies on colorectal cancer cells (SW480 and SW620) have indicated that this compound treatment can significantly reduce proliferation and induce an arrest in the G1 phase of the cell cycle alzdiscovery.org. In these cells, this compound also promoted apoptosis alzdiscovery.org.
In gastric cancer cell lines (MKN45, MKN74, and KATOIII), exposure to this compound effectively inhibited cell proliferation nih.gov. Furthermore, in a mouse model of gastric cancer, CCR5 antagonism modulated the expression of genes known for their role in cancer growth, including those related to the cell cycle alzdiscovery.orgnih.gov.
Research in classical Hodgkin lymphoma (cHL) cells has shown that this compound treatment can slightly slow cell growth and slightly increase the percentage of cells in the G1 phase of the cell cycle researchgate.net. While it influenced the cell cycle distribution, in this specific context, this compound treatment did not induce apoptosis or necrosis as measured by certain markers researchgate.net.
Studies investigating the effects of CCR5 blockade by this compound on cell cycle-related signaling cascades in colorectal cancer cells (SW620) have revealed alterations in the expression of numerous genes involved in cell cycle regulation, including cyclins and cyclin-dependent kinases (CDKs) aacrjournals.orgresearchgate.net. This suggests that CCR5 antagonism can interfere with the molecular machinery controlling cell cycle progression researchgate.net.
The impact of this compound on cell cycle regulation appears to be cell type-specific and context-dependent, with notable effects observed in various cancer models, contributing to reduced proliferation and, in some cases, increased apoptosis.
Here is a summary of research findings on this compound's impact on cell cycle regulation in specific cell types:
| Cell Type | Observed Effect of this compound on Cell Cycle | Key Findings | Source |
| Colorectal Cancer (SW480, SW620) | Reduced proliferation, G1 phase arrest, induction of apoptosis. | Significantly reduced proliferation, induced G1 arrest, promoted apoptosis, increased cleaved caspases. | alzdiscovery.org |
| Gastric Cancer (MKN45, MKN74, KATOIII) | Inhibition of cell proliferation. | Effectively inhibited cell proliferation in vitro. Modulated expression of cancer growth-related genes in vivo. | alzdiscovery.orgnih.gov |
| Classical Hodgkin Lymphoma (cHL) | Slightly slowed cell growth, slight increase in G1 phase percentage. | Slightly increased percentage of cells in G1 phase; did not induce apoptosis/necrosis in this context. | researchgate.net |
| Colorectal Cancer (SW620) | Interference with cell cycle-related signaling cascades. | Altered expression of genes involved in cell cycle regulation (cyclins, CDKs). | aacrjournals.orgresearchgate.net |
Broader Therapeutic Potential of Ccr5 Antagonism Beyond Hiv-1
CCR5 in Cancer Progression and Therapy Research
CCR5 is expressed in various cancers, and its expression level has been suggested as a negative prognostic marker in some malignancies. nih.govresearchgate.net The interaction between cancer cells and their microenvironment, often mediated by chemokine axes like CCR5/CCL5, is critical for tumor progression, including invasion and metastasis. researchgate.net Maraviroc, as a CCR5 antagonist, has been investigated for its potential to interfere with these processes. nih.govresearchgate.net
Anti-Tumor Activity and Apoptosis Induction in Cancer Cell Lines
Studies have explored the direct effects of this compound on cancer cell lines, observing potential anti-tumor activity and the induction of apoptosis. For instance, in vitro experiments with human gastric cancer cell lines (MKN45, MKN74, and KATOIII) demonstrated that exposure to this compound effectively inhibited cell proliferation. nih.govresearchgate.netresearchgate.net Additionally, this compound has been shown to induce apoptosis in colorectal cancer cells, leading to a significant arrest in the G1 phase of the cell cycle and observable morphological signs of apoptosis. researchgate.net Analysis at the mRNA and protein levels revealed significant modulation of multiple apoptosis-relevant genes and increases in cleaved caspases in these cells upon this compound exposure. researchgate.net In acute lymphoblastic leukemia (ALL) cell lines, this compound significantly inhibited cell survival and induced apoptosis, with TUNEL staining showing a notable increase in apoptotic cells after treatment. nih.gov
Here is a summary of selected in vitro findings on this compound's effects on cancer cell lines:
| Cancer Type | Cell Lines Tested | Observed Effects (In Vitro) | Source(s) |
| Gastric Cancer | MKN45, MKN74, KATOIII | Inhibited cell proliferation. nih.govresearchgate.netresearchgate.net | nih.govresearchgate.netresearchgate.net |
| Colorectal Cancer | SW480, SW620 | Induced cytotoxicity and apoptosis, G1 cell cycle arrest. researchgate.net | researchgate.net |
| Acute Lymphoblastic Leukemia | MOLT-3, TALL-104, SUP-B15 | Inhibited survival, induced apoptosis. nih.gov | nih.gov |
| Breast Cancer | MDA-MB-231 | Enhanced cell killing by DNA-damaging agents. aacrjournals.orgnih.gov | aacrjournals.orgnih.gov |
| Hodgkin Lymphoma | L-1236, L-428, KM-H2, HDLM-2 | Decreased cell growth, synergized with chemotherapy. haematologica.orghaematologica.org | haematologica.orghaematologica.org |
Modulation of Cancer Cell Migration and Adhesion
CCR5 and its ligands are involved in the migration and adhesion of cancer cells, processes crucial for metastasis. nih.govresearchgate.net In vitro experiments have shown that CCR5 antagonism by this compound reduces the migration of gastric cancer cells induced by CCR5 ligands such as macrophage inflammatory protein 1α (MIP-1α), MIP-1β, and RANTES. nih.govresearchgate.net this compound also effectively inhibited the adhesion of these gastric cancer cell lines to the murine peritoneum. nih.govresearchgate.netresearchgate.net In breast cancer models, CCR5 inhibition by this compound or vicriviroc reduced in vitro invasion of basal breast cancer cells without affecting cell proliferation or viability. aacrjournals.org Furthermore, this compound significantly inhibited the migration and adhesion of ALL cells in response to relevant stimuli, including chemotaxis towards CXCL12 and CXCL13. nih.gov This suggests a potential mechanism by which this compound could interfere with the dissemination of cancer cells. nih.gov
Here is a summary of selected in vitro findings on this compound's effects on cancer cell migration and adhesion:
| Cancer Type | Cell Lines Tested | Observed Effects (In Vitro) | Source(s) |
| Gastric Cancer | MKN45, MKN74, KATOIII | Reduced migration induced by MIP-1α, MIP-1β, and RANTES; inhibited adhesion to peritoneum. nih.govresearchgate.netresearchgate.net | nih.govresearchgate.netresearchgate.net |
| Breast Cancer | MDA-MB-231 | Reduced invasion. aacrjournals.org | aacrjournals.org |
| Acute Lymphoblastic Leukemia | SUP-B15 | Inhibited migration and adhesion; inhibited chemotaxis towards CXCL12 and CXCL13. nih.gov | nih.gov |
Preclinical in vivo studies using mouse models have also provided evidence for this compound's ability to reduce cancer burden and metastasis. Administration of this compound effectively reduced the extent of peritoneal disease and increased survival in mice inoculated with gastric cancer cells. nih.govresearchgate.net this compound treatment also reduced tumor burden in xenograft models of gastric cancer. nih.govresearchgate.netresearchgate.net In models of breast cancer, CCR5 antagonists like this compound and vicriviroc blocked metastasis of human breast cancer xenografts by inhibiting homing. aacrjournals.orgnih.gov this compound has also been shown to reduce prostate cancer cell metastasis to various organs in immunocompetent mice. aacrjournals.orgnih.gov In a mouse model of colorectal cancer, this compound suppressed tumor formation and reduced the accumulation of cancer-associated fibroblasts. oncotarget.com
CCR5 in Inflammatory Diseases Research
CCR5 plays a significant role in inflammatory responses by directing immune cell migration to sites of inflammation. frontiersin.orgtandfonline.com Dysregulation of this process is implicated in various inflammatory diseases. frontiersin.orgtandfonline.com this compound's ability to block CCR5 has led to investigations into its potential to attenuate inflammation in different disease contexts. frontiersin.orgtandfonline.com
Attenuation of Inflammation (e.g., Sepsis-Associated Liver Injury, Lung Inflammation)
Studies have explored the anti-inflammatory effects of this compound in models of inflammatory diseases. In a rat model of trauma-hemorrhage-induced hepatic injury, this compound administration attenuated the injury and decreased cytokine production. nih.gov This suggests a role for this compound in regulating hepatic injury following such events. nih.gov this compound has also shown potential in alleviating sepsis-associated liver injury (SALI) in a CLP (cecal ligation and puncture) model, significantly ameliorating liver damage and reducing inflammatory cytokine concentrations within liver tissue. nih.govdntb.gov.uaresearchgate.net
In models of lung inflammation, this compound has demonstrated beneficial effects. In mice exposed to cigarette smoke and infected with influenza A virus, this compound treatment significantly improved lung function deterioration and survival rate, while also reducing the accumulation of neutrophils and macrophages in lung tissue. mdpi.com this compound pre-treatment attenuated LPS-mediated acute lung injury in mice, reducing alveolar septal thickening and inflammatory cell infiltration. imrpress.com This was accompanied by decreased pulmonary edema, reduced cellularity and total protein in bronchoalveolar lavage fluid, and suppressed pulmonary expression and serum levels of pro-inflammatory cytokines. imrpress.com
Regulation of Inflammatory Cell Recruitment and Infiltration
A key mechanism by which CCR5 contributes to inflammation is by mediating the recruitment and infiltration of inflammatory cells. frontiersin.orgtandfonline.com this compound, as a CCR5 antagonist, interferes with this process. In a mouse model of genetic dyslipidemia, this compound reduced atherosclerotic progression by interfering with inflammatory cell recruitment into plaques. ahajournals.orgahajournals.org In mice with ritonavir-induced inflammation, this compound reduced plaque areas and macrophage infiltration, and downregulated the expression of adhesion molecules and chemokines involved in inflammatory cell recruitment. ahajournals.orgnih.gov
In the context of lung inflammation, this compound treatment reduced neutrophil and macrophage infiltration in the airways and lung tissue of mice exposed to cigarette smoke and infected with influenza A virus. mdpi.com This effect on neutrophil accumulation might be indirectly related to this compound's effect on macrophages, which produce neutrophil chemoattractants. mdpi.com this compound also reduced monocyte migration towards the site of injury and macrophage accumulation in the lungs, consistent with the known role of CCR5 in monocyte chemotaxis. mdpi.com
Role of Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) Pathway Modulation
Research suggests that the anti-inflammatory effects of this compound in certain contexts may involve the modulation of the peroxisome proliferator-activated receptor gamma (PPARγ) pathway. nih.govresearchgate.netresearchgate.net The PPARγ pathway is known to exert anti-inflammatory effects. nih.govresearchgate.netresearchgate.net Studies investigating trauma-hemorrhage-induced hepatic and lung injury in rats have explored whether this compound's protective effects are mediated through a PPARγ-dependent pathway. nih.govresearchgate.netresearchgate.net Results from these studies suggest that PPARγ might play a key role in this compound-mediated protection against hepatic and lung injury following trauma-hemorrhage. nih.govresearchgate.netresearchgate.net Coadministration of a PPARγ antagonist with this compound abolished the beneficial effects of this compound on inflammatory parameters and organ injury in these models. nih.govresearchgate.net
Relevance in Atherosclerosis Research
Atherosclerosis, a chronic inflammatory disease of the arteries, involves the recruitment of immune cells to the arterial wall, contributing to plaque formation and progression. medsci.org Evidence suggests a significant role for CCR5 and its ligands, such as CCL3, CCL4, and CCL5 (RANTES), in the initiation and progression of atherosclerosis. ahajournals.orgmedsci.orgnih.gov CCR5 is expressed on various cell types involved in atherosclerosis, including endothelial cells, monocytes, and macrophages. medsci.orgfrontiersin.org Activation of CCR5 by its ligands can promote inflammation and the release of cytokines that contribute to plaque formation and instability. medsci.org
Studies in murine models have demonstrated that CCR5 deficiency or antagonism can reduce the development and progression of atherosclerosis. ahajournals.orgnih.govnatap.org For instance, in apolipoprotein E-deficient (ApoE−/−) mice, a model prone to atherosclerosis, CCR5 deficiency has shown a protective effect against diet-induced atherosclerosis and reduced infiltration of mononuclear and Th1 type immune cells. frontiersin.org this compound, as a CCR5 antagonist, has been investigated for its potential anti-atherogenic effects. In murine models, this compound has been shown to reduce atherosclerotic progression by modulating inflammatory cell recruitment to plaques and systemic inflammation. ahajournals.orgnih.gov Specifically, this compound treatment in ApoE−/− mice inhibited plaque development and reduced macrophage infiltration. ahajournals.orgnih.gov It also downregulated the local expression of adhesion molecules like vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1), as well as chemokines like RANTES and IL-2 within the plaques. ahajournals.orgnih.gov
Clinical studies exploring the effect of this compound on atherosclerosis in humans are limited but suggest potential benefits. A pilot study in HIV-positive patients at high cardiovascular risk showed that this compound treatment improved markers of cardiovascular risk, endothelial dysfunction, arterial stiffness, and early carotid atherosclerosis. alzdiscovery.org These improvements included significant enhancements in brachial flow-mediated dilation, carotid-femoral pulse wave velocity, and carotid intima-media thickness. alzdiscovery.org
The role of the CCL5/RANTES-CCR5 axis is considered pivotal in the development and progression of atherosclerotic inflammatory disease. ahajournals.org CCL5 is detected in atherosclerotic plaques and is often upregulated in later stages of the disease. ahajournals.org Released by activated platelets, CCL5 can trigger the accumulation of monocytes on inflamed endothelium. ahajournals.org CCR5 also influences the recruitment of endothelial progenitor cells (EPCs) to plaques, which are important for maintaining a functional endothelium and have been shown to promote plaque regression. frontiersin.orgemjreviews.com CCR5 overexpression in mice has been linked to increased EPC recruitment to lesions and improved endothelial function. emjreviews.com
Data from studies on CCR5 antagonism in atherosclerosis:
| Study Type | Model/Cohort | Intervention | Key Findings | Citation |
| Murine | ApoE−/− mice (ritonavir-induced atherogenesis) | This compound | Reduced plaque areas, macrophage infiltration, downregulated ICAM-1, VCAM-1, RANTES, IL-2 expression in plaques. Reversed ritonavir-induced proinflammatory profile. | ahajournals.orgnih.gov |
| Murine | ApoE−/− mice (late-stage atherosclerosis) | This compound | Inhibited progression of spontaneous atherosclerosis, reduced macrophage infiltration, lowered expression of adhesion molecules and RANTES in plaques. | ahajournals.orgnih.gov |
| Human | HIV-positive, high CV risk (pilot study) | This compound (300 mg/day for 24 weeks) | Improved brachial flow-mediated dilation, carotid-femoral pulse wave velocity, carotid intima-media thickness. | alzdiscovery.org |
| Murine | ApoE−/− mice | CCR5 deficiency or antagonism (general) | Reduced atherosclerotic burden, reduced systemic secretion of proinflammatory Th1 cytokines. | ahajournals.orgnih.gov |
Potential in Other Immunological Conditions
Beyond atherosclerosis, the role of CCR5 in immune cell trafficking and inflammation has led to investigations into the potential of CCR5 antagonism in various other immunological and inflammatory conditions. frontiersin.orgtandfonline.comresearchgate.net CCR5 is expressed on key immune cells, including T cells, macrophages, and dendritic cells, and its activation mediates immune responses by binding chemokines like CCL3, CCL4, and CCL5. researchgate.net In chronic inflammatory diseases, affected sites often show increased inflammatory cell infiltrates expressing CCR5 and high levels of its ligands. tandfonline.comnih.gov
The therapeutic potential of targeting CCR5 in inflammatory diseases stems from the observation that blocking CCR5 is likely to specifically affect infiltrating cells at inflammatory sites, potentially leading to reduced local inflammation with minimal systemic immunosuppression. tandfonline.com
Research has explored CCR5 antagonism in conditions such as:
Inflammatory Bowel Disease (IBD): CCR5 is involved in the pathogenesis of IBD and is highly expressed in the colonic mucosa of patients with the condition. archivesofmedicalscience.com Studies using CCR5 antagonist peptides in rat models of colitis have shown inhibitory effects on inflammatory cell infiltration. archivesofmedicalscience.com
Rheumatoid Arthritis (RA): CCR5 is abundantly expressed in the synovium and inflammatory infiltrates in RA. nih.gov While initial clinical trials with CCR5 antagonists, including this compound, in RA patients showed no significant clinical efficacy, the complex nature of RA and the chemokine system warrant further investigation into patient selection and dosing strategies. nih.gov
Multiple Sclerosis (MS): CCR5 is expressed on immune cells within inflammatory lesions in MS and may contribute to their recruitment and activation in the inflamed tissue. frontiersin.org Evidence suggests that while the CCR5Δ32 allele (associated with non-functional CCR5) is not linked to MS risk, the disease may be less severe in carriers, implying that CCR5 antagonists might reduce disease activity. frontiersin.org Studies in experimental autoimmune encephalomyelitis (EAE), a mouse model of MS, have shown that CCR5 knockout or antagonism can reduce infiltrating immune cells in the central nervous system (CNS). mdpi.com
Graft-versus-Host Disease (GvHD): The CCR5 receptor appears to play a central role in modulating immune cell trafficking to sites of inflammation, which is crucial in the development of acute GvHD. vax-before-travel.com Clinical studies support the concept that blocking CCR5 can reduce the clinical impact of acute GvHD without significantly affecting bone marrow stem cell engraftment. vax-before-travel.com this compound treatment has been reported to increase CCR5 expression on T cells and reduce T cell activation in peripheral blood, suggesting a potential protective effect against GvHD through modulation of allo-reactive donor T cell responses. frontiersin.org
Sarcoidosis: CCR5 is involved in the TH1 signaling cascade implicated in sarcoidosis. jiaci.org A case report described an HIV-infected patient with pulmonary sarcoidosis who showed both immune restoration and relief of respiratory symptoms following intensification of antiretroviral therapy with this compound. jiaci.org This observation suggests that this compound, as a selective CCR5 inhibitor, warrants further evaluation in sarcoidosis, potentially in non-HIV-infected patients as well. jiaci.org
Neuroinflammatory Diseases: Chemokine receptors, including CCR5, are implicated in various CNS inflammatory diseases and play roles in the recruitment and positioning of immune cells within tissues. researchgate.net this compound has been hypothesized to offer neuroprotective benefits in conditions where CCR5 promotes deleterious neuroinflammation, particularly those involving CD8+ T cells. researchgate.net This includes conditions like HIV-associated neurocognitive disorders. researchgate.net
The involvement of CCR5 in a broad array of chronic inflammatory diseases positions CCR5 antagonism as a potential therapeutic strategy. frontiersin.org While this compound is the only CCR5 antagonist currently approved for clinical use (in HIV), research continues to explore its potential and the potential of other CCR5-targeting approaches in various inflammatory and immunological conditions. frontiersin.orgresearchgate.net
Table of CCR5 Antagonism Research in Other Immunological Conditions:
| Condition | Relevance of CCR5 | Research Findings / Potential Benefit | Citation |
| Inflammatory Bowel Disease (IBD) | Involved in pathogenesis, highly expressed in colonic mucosa. | CCR5 antagonist peptides reduced inflammatory cell infiltration in rat models of colitis. | archivesofmedicalscience.com |
| Rheumatoid Arthritis (RA) | Abundantly expressed in synovium and inflammatory infiltrates. | Initial clinical trials with this compound and other antagonists showed no significant efficacy, but further investigation is considered. | nih.gov |
| Multiple Sclerosis (MS) | Expressed on immune cells in inflammatory lesions, contributes to recruitment. | CCR5 knockout or antagonism reduced infiltrating immune cells in EAE mouse model. Potential to reduce disease activity. | frontiersin.orgmdpi.com |
| Graft-versus-Host Disease (GvHD) | Central role in immune cell trafficking to inflammatory sites. | Blocking CCR5 reduced clinical impact of acute GvHD in studies. This compound modulated T cell responses. | frontiersin.orgvax-before-travel.com |
| Sarcoidosis | Involved in TH1 signaling cascade. | Case report showed immune restoration and symptom relief with this compound in an HIV-infected patient. Potential for evaluation in non-HIV patients. | jiaci.org |
| Neuroinflammatory Diseases | Implicated in CNS inflammation, roles in immune cell recruitment. | Hypothesized neuroprotective benefits in conditions with deleterious neuroinflammation, including HIV-associated neurocognitive disorders. | researchgate.net |
Future Directions in Maraviroc Academic Research
Development of Novel Maraviroc-Related Analogues with Improved Pharmacological Profiles
Research is ongoing to design and synthesize novel compounds structurally related to this compound with the aim of achieving improved pharmacological profiles. This includes efforts to enhance potency, improve pharmacokinetic properties, and potentially overcome existing limitations such as susceptibility to resistance or drug interactions. Computational studies, including molecular docking and dynamics simulations, are being employed to guide the design of these new analogues by predicting their binding interactions with the CCR5 receptor and evaluating their stability. researchgate.netnih.govtandfonline.com For instance, in silico investigations have explored novel this compound analogues for potential dual inhibition of CCR5 and other targets like SARS-CoV-2 Mpro, suggesting structures that could be synthesized and evaluated in vitro and in vivo. researchgate.nettandfonline.com
Deeper Elucidation of Resistance Mechanisms and Predictive Markers
Understanding the mechanisms by which HIV-1 develops resistance to this compound is crucial for its continued effectiveness and the development of next-generation CCR5 antagonists. Research indicates that resistance can arise through mutations in the V3 loop of the viral gp120 protein, allowing the virus to utilize this compound-bound CCR5 coreceptors. oup.comnih.gov Another mechanism involves a shift in viral tropism from CCR5 to CXCR4 usage. oup.comnih.gov Future research aims to more deeply elucidate these resistance pathways and identify predictive markers that can forecast treatment failure. Studies have utilized phenotypic assays, such as the PhenoSense Entry assay, to identify reduced maximal percentage inhibition (MPI) as a marker of this compound resistance in patient samples, correlating this with V3 loop mutations. natap.org However, the diverse patterns of amino acid changes observed suggest the need for further research to establish canonical mutations and develop reliable genotypic algorithms for predicting resistance. nih.gov Research is also exploring the predictive value of immunological markers, such as the CD4/CD8 ratio, in identifying patients at higher risk of disease progression despite viral suppression, which could potentially inform treatment strategies including the use of this compound. clinicalstudydatarequest.com
Advanced Computational and Structural Studies for Rational Drug Design
Computational and structural studies play a vital role in the rational design of improved CCR5 inhibitors. The availability of the crystal structure of the CCR5-Maraviroc complex has significantly advanced the ability to investigate protein-ligand interactions at a molecular level. scirp.org Techniques such as molecular dynamics (MD) simulations, accelerated MD simulations, and free energy calculations are employed to understand the detailed interaction mechanisms between this compound and CCR5. nih.gov These studies provide insights into the flexibility of different parts of the this compound molecule within the binding pocket and the energetic landscape of its dissociation. nih.gov This information is invaluable for designing more potent and effective CCR5 antagonists with desired binding characteristics and improved pharmacological profiles. nih.govscirp.org Structure-based pharmacophore modeling, using this compound as a template, is also utilized for virtual screening of large databases to identify novel CCR5 inhibitors with favorable properties. scirp.org
Retrosynthesis Analysis
AI-Powered Synthesis Planning: Our tool employs the Template_relevance Pistachio, Template_relevance Bkms_metabolic, Template_relevance Pistachio_ringbreaker, Template_relevance Reaxys, Template_relevance Reaxys_biocatalysis model, leveraging a vast database of chemical reactions to predict feasible synthetic routes.
One-Step Synthesis Focus: Specifically designed for one-step synthesis, it provides concise and direct routes for your target compounds, streamlining the synthesis process.
Accurate Predictions: Utilizing the extensive PISTACHIO, BKMS_METABOLIC, PISTACHIO_RINGBREAKER, REAXYS, REAXYS_BIOCATALYSIS database, our tool offers high-accuracy predictions, reflecting the latest in chemical research and data.
Strategy Settings
| Precursor scoring | Relevance Heuristic |
|---|---|
| Min. plausibility | 0.01 |
| Model | Template_relevance |
| Template Set | Pistachio/Bkms_metabolic/Pistachio_ringbreaker/Reaxys/Reaxys_biocatalysis |
| Top-N result to add to graph | 6 |
Feasible Synthetic Routes
Featured Recommendations
| Most viewed | ||
|---|---|---|
| Most popular with customers |
Disclaimer and Information on In-Vitro Research Products
Please be aware that all articles and product information presented on BenchChem are intended solely for informational purposes. The products available for purchase on BenchChem are specifically designed for in-vitro studies, which are conducted outside of living organisms. In-vitro studies, derived from the Latin term "in glass," involve experiments performed in controlled laboratory settings using cells or tissues. It is important to note that these products are not categorized as medicines or drugs, and they have not received approval from the FDA for the prevention, treatment, or cure of any medical condition, ailment, or disease. We must emphasize that any form of bodily introduction of these products into humans or animals is strictly prohibited by law. It is essential to adhere to these guidelines to ensure compliance with legal and ethical standards in research and experimentation.


